Poster: GunnVistaPingry_US

iGEM Student Team Members: Navya Lam (Team Lead), Aaron Chacko Mundanilkunathil, Alisha Saboowala, Charuthi Arul, Sanjana Biswas, Stephanie Muggler, Dia Dhariwal, Aditi Sadwelkar, Dorothy Lam
Advisors: Dr. Samuel Perli, Siva Palagummi, Craig M. Trester, Lauren Turetsky
Abstract
CRISPRi is a powerful tool for modulating gene expression in human cells. By designing a gRNA homologous to the target gene of interest, one can achieve targeted knockdown of the specific gene of interest. However, with current methodologies, one has to screen multiple gRNA sequences for efficient targeting while minimizing off-target effects. We present a prediction model for identifying the best gRNA sequence for efficient gene targeting in human cells. Starting with experimental data from knocking down specific genes using several gRNAs in iPS cells, we leverage machine learning to inform better selection of the gRNA. Our tool will be invaluable for designing gene targeting gRNAs and will reveal underlying biochemical principles governing CRISPR efficiency.

- Generate improved AI/ML model to predict quality sgRNA
- Simple Interface
- Off-target stringency
- Support other hosts and different algorithms

Wet-lab experience, Shinya Yamanaka, CRISPR pioneer Jennifer Doudna, State-of-the-art creator Jonathan S. Weissman

Before iGEM, our team lead Navya worked in the Yamanaka Lab in Gladstone Institutes. Using CRISPR technology, she researched the role of a particular protein in the behavior of paraspeckles, subnuclear organelles in both mice and humans. During her research, she ran into a major wet lab setback and found that using software solutions was incredibly effective for transcending obstacles. The laborious and expensive lab work also required testing sgRNA strands in live cells to find a good fit. The unwieldiness of this testing process inspired Navya, her supervisor Dr. Perli, and the rest of the iGEM team to study computational solutions for sgRNA selection. We were fascinated by the efficiency of artificial intelligence as it dramatically streamlined an unwieldy biological process that would ordinarily have been done through lengthy trial and error.


CRISPR guide RNA selection: arduous, outdated and expensive
CRISPRi (dcas9-KRAB), an inactive variant of CRISPR, preempts gene expression by blocking RNA transcription, yielding low relative gene expression. However, putting CRISPRi into a cell requires an effective guide RNA. The state-of-the-art for computerized guide RNA efficacy prediction, the Weissman Algorithm, ranks the guide RNAs. When inserting the top ten guide RNAs as predicted by the algorithm, the results are as shown

Given the unpredictability shown above in guide RNA ranking with the Weissman algorithm, it would be wisest for one to test all the ten guide RNAs to see which was most effective at its duty. Moreover, the designing, inserting and screening of CRISPR is an arduous, expensive and time consuming process. This unreliability in guide RNA is also a large roadblock to utilizing CRISPR in therapeutics.

One drawback of the Weissman algorithm was that it used phenotypic data to determine the efficacy of a guide RNA in training its algorithm.
Our team found a solution through Real-Time qRT-PCR to quantitatively measure CRISPRi silencing, a method not available to Weissman when he created the algorithm.

Therefore, the wet lab Real-Time qRT-PCR data measuring sgRNA effectiveness could be overlaid on the existing algorithm to train it. Then, the algorithm could be improved by various other tweaks so a better guide RNA efficacy predictor would result.

 Odigos Model Generator: Generates AI/ML based model that can be used to predict guide RNA score that can be used with CRISPRi/Cas systems.
Odigos Model Generator: Generates AI/ML based model that can be used to predict guide RNA score that can be used with CRISPRi/Cas systems.Odigos Guide Predictor: Outputs sgRNAs that can be used with CRISPRi/CAS for any gene of interest using the model generated by the Model Generator. It also considers off-target stringency as well in suggesting quality guides.
Our software is based on Python 3.8 modules and end user interfaces are exposed as Jupyter Notebooks.
Third Party Packages:
ViennaRNA: The ViennaRNA Package is a set of standalone programs and libraries used for prediction and analysis of RNA secondary structures. It is maintained and managed by Theoretical Biochemistry group within the Institute for Theoretical Chemistry which in turn is part of University of Vienna.
Bowtie: It is an ultrafast, memory-efficient short read aligner geared toward quickly aligning large sets of short DNA sequences (reads) to large genomes.
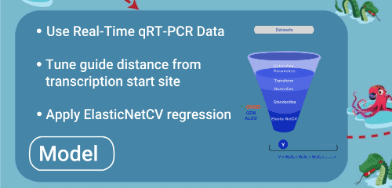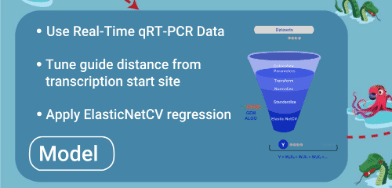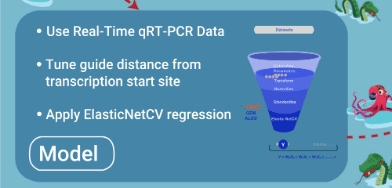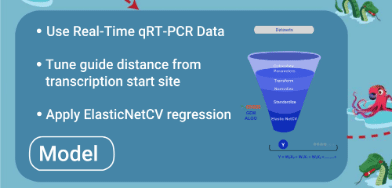


- Model is trained with quality qRT-PCR data
- Model is tuned to have square of distance of guide from primary/secondary tss as additional feature
- Best model is selected after source data is randomly divided into different training and test records
y1 = w1x1 + w2x2 + w3x3 + w4x4 + w5x5 + w6x6 + w7x7 + w8x8 + w9x9 + w10x10 + w11x11 + w12X12 + w13x13
y1 -- Score of the Guide
w1 ... w13 -- Weights of features contributing to score
x1 … x13 -- Features contributing to score as predicted by algorithm.
 Distance -- Distance of the Guide from Primary/Secondary TSS
Distance -- Distance of the Guide from Primary/Secondary TSSHomopolymers -- Number of nucleotides in the sequence
Base Table -C -- Existence of nucleotide ‘C’ at distance of 10 base pairs from PAM
Base Dimers -- Existence of specified dimer from specified distance from PAM


- Odigos Predictor uses the model trained by Odigos Model Generator
- Uses the off-target stringency in suggesting quality sgRNAs
Off-target stringency is based on phred-quality score and uses bowtie to calculate quality guides with different thresholds. Following off-target levels are considered.
offTargetLevels = [['31_nearTSS', '21_genome'],['31_nearTSS'],['21_genome'],['31_2_nearTSS'],['31_3_nearTSS']]
Highest stringency to lowest are in the following order.
- Only one match in 500bp flank region with quality threshold 31 and entire genome with quality threshold 21
- Only one match in the 500bp flanking region
- Only one match in the entire genome with 21 quality threshold value
- Only two matches in the 500bp flanking region with 31 threshold value
- Only three matches in the 500bp flanking region with 31 threshold value

 USCF Yamanaka Lab
USCF Yamanaka Lab
Dr. Perli used the guides suggested by Weissman Machine learning algorithm using phenotype based scoring and off-target filtering to do Real-Time qRT-PCR experiments. And found few guides hitting the target better compared to others. This itself proves that a better Machine Learning based algorithm that is trained with quality data would predict guides better. When we used our algorithm to predict scores of the guides they are very close to lab results compared to Weissman lab scores.
NYC_Earthians
NYC Earthians another iGEM 2020 competing team was doing a project based on CRISPR targeting APOL3 gene in COW (Bovine).
When they tried to find guide RNAs they ended up with 100 odd guides and didn't know what to use and which one would be effective.
They in fact approached us to see if our tool can help them to find the right guide RNA. Our tool which can find good guides based on our Machine Learning Model including Off-target stringency is the right choice. We are in the process of adding support to Bovine so we could not give them a guide right away. This itself is another proof of our concept, how finding a good sgRNA based on ML/AI for different CRISPR systems can help biologists. We will continue adding support and will let them know once we finish testing.




 Comparison Table: Comparison table of deviance of top ten guides of different genes
Comparison Table: Comparison table of deviance of top ten guides of different genes
- The top 10 sgRNA scores predicted by our model are close to lab values for most of the genes
- Only for FMR1 and GNB2L1 genes weissman's scores are closer to lab scores compared to Odigos predictor
- EIF4G1 (gene used in study) Guides: Table containing 10 guides along with scores and off-target stringency


 Proposed End Users:
Proposed End Users:- Future iGEMers
- Additional qRT-PCR guide RNA data results of knockdowns to be added by future iGEM teams and biologists to train algorithm, a simple but effective update to a previous iGEM team’s work
- Bioengineers
- COVID-19 Investigation and Treatment
- Stem Cell Research
- Biopharma and Therapeutics



CRISPR application areas:

CRISPR types of editing:


[2]Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013 Feb 28;152(5):1173-83.
[3]Nolan, Tania, et al. “Quantification of MRNA Using Real-Time RT-PCR.” Nature News, Nature Publishing Group, 9 Nov. 2006, www.nature.com/articles/nprot.2006.236.
[4]Lorenz, Ronny and Bernhart, Stephan H. and Höner zu Siederdissen, Christian and Tafer, Hakim and Flamm, Christoph and Stadler, Peter F. and Hofacker, Ivo L.
ViennaRNA Package 2.0
Algorithms for Molecular Biology, 6:1 26, 2011, doi:10.1186/1748-7188-6-26
[5] Bowtie 1.3.0: Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome Genome Biol 10:R25.

Team members attend
- Henry M. Gunn High School, Palo Alto, CA
- Monta Vista High School, Cupertino, CA
- The Pingry School, Somerset County, New Jersey
Navya Lam: Formed and led the project team, taught necessary content, implemented human practices and collaborations, worked on the algorithm.
Aaron Chacko Mundanilkunathil: Worked on the algorithm by studying the Weissman Lab files, normalizing the scores, debugging the code, and exploring the training sets
Alisha Saboowala: Focused on improving the model through graphical representations of the variables and by debugging the code
Chauthi Arul: Worked on exploring the UCSC genome browser for protein information and doing the voiceover for team videos
Sanjana Biswas: Surveyed the literature on CRISPR and explored other biological aspects to improve our model
Stephanie Muggler: Explored ViennaRNA to analyze secondary structure impact, led in design and animation
Dia Dhariwal: Worked on outreach, validating the model and exploring the use of polynomial regressions
Dorothy Lam: Worked on website design and researching the background on the relevant genes
Dr. Samuel David Perli: Principal Investigator for the team, helped in explanation of biological concepts to team members. Introduced and explained variables that affect sgRNA accuracy reviewed the algorithm and made suggestions
Yamanaka Lab in Gladstone Institute, UCSF: Provided RT qRT-PCR data for several gene knockdowns
Professor Shinya Yamanaka: Allowed Navya for an internship opportunity, which led her to develop this software project
Craig M. Trester: Already experienced iGEM once before, helped guide us
Siva Palagummi: helped in debugging the Weissman code when it was outdated, and later helped in advising and reviewing our software.
Lauren Turetsky: Helped guide us in website design and format
Human Practices/Interviews:
Dr. Vijai Singh, Associate Professor & Head Department of Biosciences at School of Science Indrashil University in Gujarat, India. He is also the author of “Genome Engineering via CRISPR-Cas9 System”. This was published in 2020 and was very relevant to our project
Sai Chitti, Ph.D student at the cancer biology lab at La Trobe Institute For Molecular Science in Melbourne, Australia
Professor Keichiro Tomoda, program manager at Gladstone Institutes, UCSF and former PI at Center for iPS Cell Research and Applications from Center for iPS Cell Research and Application in Kyoto University, Japan
Sponsors:


