Presented by Team Warwick 2020
Tudor Onose, Jade Oh, Valeriia Nadmitova, Adam Jones, Martin Costa, Adriana Lopez Gonzalez, Aryan Gupta, Grace Goater, Andris Gyori
Abstract:
Colorectal cancer (CRC) is a leading cause of cancer death and is the 4th most common form of cancer in the UK. CRC mortality can be reduced through early intervention, so we wanted to produce a sensor that could be used to identify high risk individuals. The presence of colibactin-producing E.coli strains within the gut microbiota has been shown to contribute to host DNA damage and is found more commonly in CRC patients. Using the current literature, we found a stable by-product of colibactin production known as, N-myristoyl-D-asparagine, and modified a bacterial receptor called MmfR to detect our ligand. Unfortunately, due to the ligand’s long acyl chain, we were unable to create a sensor for N-myristoyl-D-asparagine, however we were successful in producing a N-butyryl-D-asparagine that could be evolved into a N-myristoyl-D-asparagine biosensor in future work.

Colibactin Detection: The Race for Early Colorectal Cancer Detection.

Inspiration: Colorectal Cancer
Colorectal cancers (CRC) often begin in the mucosa tissues of the colon or rectum, spreading outwards into intestine wall, and in later stages, to blood and lymph vessels that propagate the spread of the cancer to other parts of the body.(1) CRC is a very common cancer in the UK, accounting for 42 thousand cases per year.

As seen in the statistics above, mortality increases the later a cancer is discovered. The UK has below average survival rates of CRC in comparison to European averages and early detection is a vital part of reducing the number of deaths (2).

As seen in the statistics above, mortality increases the later a cancer is discovered. The UK has below average survival rates of CRC in comparison to European averages and early detection is a vital part of reducing the number of deaths (2).

Introduction: Colibactin
Gut microbiota also plays a role in the development of CRC. Bacteria such as E.coli can possess a genomic ‘pks’ island, containing the enzymatic machinery to produce colibactin. Colibactin (seen below) is a highly reactive natural product that can damage the DNA of a person’s cells through double strand breakage and acylation (3).

The presence of pks+ bacteria is associated with higher occurrence of CRC, so could be used to find high risk individuals (4).
Therefore, we want to produce a sensor that can detect the presence of this bacteria in a fast, reliable non-invasive way, so that people at a higher risk of developing CRC can receive more regular monitoring and prevent the progression of potential CRC.

The presence of pks+ bacteria is associated with higher occurrence of CRC, so could be used to find high risk individuals (4).
Therefore, we want to produce a sensor that can detect the presence of this bacteria in a fast, reliable non-invasive way, so that people at a higher risk of developing CRC can receive more regular monitoring and prevent the progression of potential CRC.
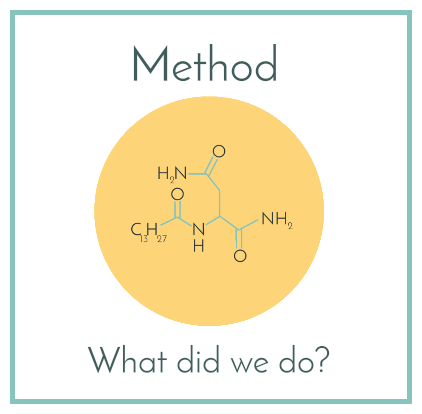
Method
Due to coronavirus, our project was limited to dry lab experiments. This is what we did:
- We investigated colibactin’s literature and found the molecule too large and too reactive to be the ligand for our biosensor.
- A small molecule that was cleaved from precolibactin in the final stage of colibactin production called N-myristoyl-D-asparagine (shown below) was determined to be a better option for our ligand. (5)
- We identified a suitable template receptor, MmfR, as it’s native ligand MMF possessed a similar structure to N-myr-D-Asn(6).
- In the dry lab, we used Rosetta and Avogandro to dock various ligands to our receptor and then modify the receptor’s amino acid sequence to better accommodate our target.


Results
The large difference in size between N-myr-D-Asn and the MMF ligand of MmfR meant we opted for a two-step approach. First we designed an N-acetyl-D-myristoyl sensor to ensure that the software could create the hydrogen bonds needed for docking of the asparagine residue and then we used the best results as design templates for our final sensor.
However, this failed as the N-myristoyl-D-asparagine's 14C alkyl chain was too long to design a binding pocket for with the template.
We later managed to design an N-butyryl-D-asparagine receptor, able to bind a version of our ligand with a shorter alkyl chain (the successful receptor design is shown below).

However, this failed as the N-myristoyl-D-asparagine's 14C alkyl chain was too long to design a binding pocket for with the template.
We later managed to design an N-butyryl-D-asparagine receptor, able to bind a version of our ligand with a shorter alkyl chain (the successful receptor design is shown below).


The Future
 We were unable to produce a receptor for N-myristoyl-D-asparagine, however, using the data we produced, a future team could gradually evolve the N-butyryl-D-asparagine into the desired ligand(7). We also experienced limitations due to COVID-19 and were unable to do any wet lab testing, but in the future, it may be possible to test our receptor using synthetic ligands.
We were unable to produce a receptor for N-myristoyl-D-asparagine, however, using the data we produced, a future team could gradually evolve the N-butyryl-D-asparagine into the desired ligand(7). We also experienced limitations due to COVID-19 and were unable to do any wet lab testing, but in the future, it may be possible to test our receptor using synthetic ligands.
Our hope for an eventual testing kit would be a cell free extract containing our receptor on a vector along with a reporter protein like GFP and necessary transcription and translation machinery. In practise, we would hope this test could be used with stool samples and produce fluorescence in the presence of colibactin-producing E.coli (as shown below).


Human Practices
 The feasibility and purpose of our research project, and our responsibilities as scientists, were at the front of our minds. Our human practice research and reflections influenced our development process - such as our proposed end-user which changed multiple times throughout the project, from healthcare practitioner, to patient, then to practitioner again as we considered feedback and learnt more about CRC screening and the issues facing patients.
The feasibility and purpose of our research project, and our responsibilities as scientists, were at the front of our minds. Our human practice research and reflections influenced our development process - such as our proposed end-user which changed multiple times throughout the project, from healthcare practitioner, to patient, then to practitioner again as we considered feedback and learnt more about CRC screening and the issues facing patients.
Our Human Practices research also informed several endeavours, such as CRC awareness campaigns, that we aim to continue pursuing after iGEM. We also believe that the information we've collected throughout the process will be helpful to future teams working on similar topics.

Conclusion
In conclusion, while we were unable to make a receptor for N-myr-D-asn, future work could produce a receptor that is able to detect the presence of pks+ E.coli based upon the foundation we laid out here, and with the addition of a reporter protein, we could produce a test that would be rapid and could be utilised by a medical practitioner to identify at risk patients for CRC.



Acknowledgements, Sponsors and References
Acknowledgements
Dr. Christophe Corre,
Dr. Emzo De Los Santos,
Patrick Capel
Megha Bawa
Marion Dugué
Chris Graham
Special thanks to:
Ms. Amanda Bishop
Dr. Nasir Rajpoot
Dr. Guha Tanaya
Dr. Nicholas Dale
Dr. Robin Allaby
Mr. Keith Chester Dacanay
Warwick Biology Society
References:
1. Cancer.org. 2020. What Is Colorectal Cancer?| How Does Colorectal Cancer Start?. [online] Available at: [Accessed 8 November 2020].
2. Cancer Research UK. 2020. Bowel Cancer Statistics. [online] Available at: [Accessed 8 November 2020].
3. Nougayrede, J., 2006. Escherichia coli Induces DNA Double-Strand Breaks in Eukaryotic Cells. Science, 313(5788), pp.848-851.
4. Arthur, J., Perez-Chanona, E., Mühlbauer, M., Tomkovich, S., Uronis, J., Fan, T., Campbell, B., Abujamel, T., Dogan, B., Rogers, A., Rhodes, J., Stintzi, A., Simpson, K., Hansen, J., Keku, T., Fodor, A. and Jobin, C., 2012. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science, 338(6103), pp.120-123.
5. Wernke, K., Xue, M., Tirla, A., Kim, C., Crawford, J. and Herzon, S., 2020. Structure and bioactivity of colibactin. Bioorganic & Medicinal Chemistry Letters, 30(15), p.127280.
6.Zhou, S., Bhukya, H., Malet, N., Harrison, P. J., Rea, D., Belousoff, M. J., Venugopal, H., Sydor, P. K., Styles, K. M., Song, L., Cryle, M. J., Alkhalaf, L. M., Fulop, V., Challis., G. L., Corre, C., 2020. Structural basis for control of antibiotic production by bacterial hormones, BioRxiv, doi:10.1101/2020.05.02.07398
7.Arnold, F., 1998. Design by Directed Evolution. Accounts of Chemical Research, 31(3), pp.125-131.
Sponsors:

Dr. Christophe Corre,
Dr. Emzo De Los Santos,
Patrick Capel
Megha Bawa
Marion Dugué
Chris Graham
Special thanks to:
Ms. Amanda Bishop
Dr. Nasir Rajpoot
Dr. Guha Tanaya
Dr. Nicholas Dale
Dr. Robin Allaby
Mr. Keith Chester Dacanay
Warwick Biology Society
References:
1. Cancer.org. 2020. What Is Colorectal Cancer?| How Does Colorectal Cancer Start?. [online] Available at:
2. Cancer Research UK. 2020. Bowel Cancer Statistics. [online] Available at:
3. Nougayrede, J., 2006. Escherichia coli Induces DNA Double-Strand Breaks in Eukaryotic Cells. Science, 313(5788), pp.848-851.
4. Arthur, J., Perez-Chanona, E., Mühlbauer, M., Tomkovich, S., Uronis, J., Fan, T., Campbell, B., Abujamel, T., Dogan, B., Rogers, A., Rhodes, J., Stintzi, A., Simpson, K., Hansen, J., Keku, T., Fodor, A. and Jobin, C., 2012. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science, 338(6103), pp.120-123.
5. Wernke, K., Xue, M., Tirla, A., Kim, C., Crawford, J. and Herzon, S., 2020. Structure and bioactivity of colibactin. Bioorganic & Medicinal Chemistry Letters, 30(15), p.127280.
6.Zhou, S., Bhukya, H., Malet, N., Harrison, P. J., Rea, D., Belousoff, M. J., Venugopal, H., Sydor, P. K., Styles, K. M., Song, L., Cryle, M. J., Alkhalaf, L. M., Fulop, V., Challis., G. L., Corre, C., 2020. Structural basis for control of antibiotic production by bacterial hormones, BioRxiv, doi:10.1101/2020.05.02.07398
7.Arnold, F., 1998. Design by Directed Evolution. Accounts of Chemical Research, 31(3), pp.125-131.
Sponsors:

