Outreach & Education
In a normal year, the Stanford iGEM team is typically only able to engage students from the Bay Area that live geographically near Stanford's campus. However this year was an exceptional year, and to match it we organized exceptional programs that reached far beyond the San Francisco Bay Area. By taking advantage of the new online format of the iGEM competition, we were able to break free of geographical barriers and engage hundreds of students across the world in our Outreach & Education programs.

We were able to engage students across at least 20 different states and 60 cities through our initiatives.
SynBio Speaker Series: Innovative, Inspiring Talks From Experts Across The Field
Our SynBio Speaker Series consisted of a series of hour-long talks hosted weekly every Thursday at 4 PM PST on Zoom, open to anybody who had the link and wanted to join. They are now currently on our YouTube channel for anyone to learn from and have received hundreds of views collectively since being posted online. The lineup includes speakers from a wide range of subfields in synthetic biology ranging from discussions on the ethics of germline editing and citizen science, to an in-depth look at systems biology and protein engineering.

Lab Webinars: A Virtual Way To Teach Lab Techniques
The webinars function as an introduction to wet and dry lab techniques that are easily accessible for beginners to understand. They were hosted weekly every Tuesday at 4 PM PST on Zoom, and are on our YouTube channel along with the SynBio Speaker Series videos. The lab webinars were planned jointly with BIOME (Biological Interdisciplinary Open Maker Environment) which is an undergraduate student organization on Stanford's campus that funds the iGEM team, and runs a biomakerspace out of the Shriram Center for Bioengineering and Chemical Engineering. These videos were done by BIOME lab managers Colin Kalicki and Alan Tong, but the summer outreach initiatives of BIOME and iGEM were organized, planned, and advertised with close coordination.
Mentorship Program: An Online Program To Enable High Schoolers To Implement Independent Research Projects
When the online format of the iGEM competition this year was announced, our team had to cancel the typically in-person High School Internship Program. However, it was still important to us to engage high school students - not just around the Bay Area, but around the country - in our outreach efforts. It was not easy designing an online program for high school students that we felt would still be meaningful to the participants, but after lots of discussion, planning, and a lengthy outreach and application process, we were able to put together an online Mentorship Program that engaged 21 high school students from across the US in online research projects.
With our High School Mentorship Program, we wanted to bring high school students their own miniature iGEM experience; an opportunity to work in teams and execute an independent research project, with the aim of solving a relevant problem in the world. As they worked through their projects - which was in the realm of protein engineering - we were able to teach them more about biology, patent law, and bioinformatics, and to take them through how to implement a research project from start to finish.
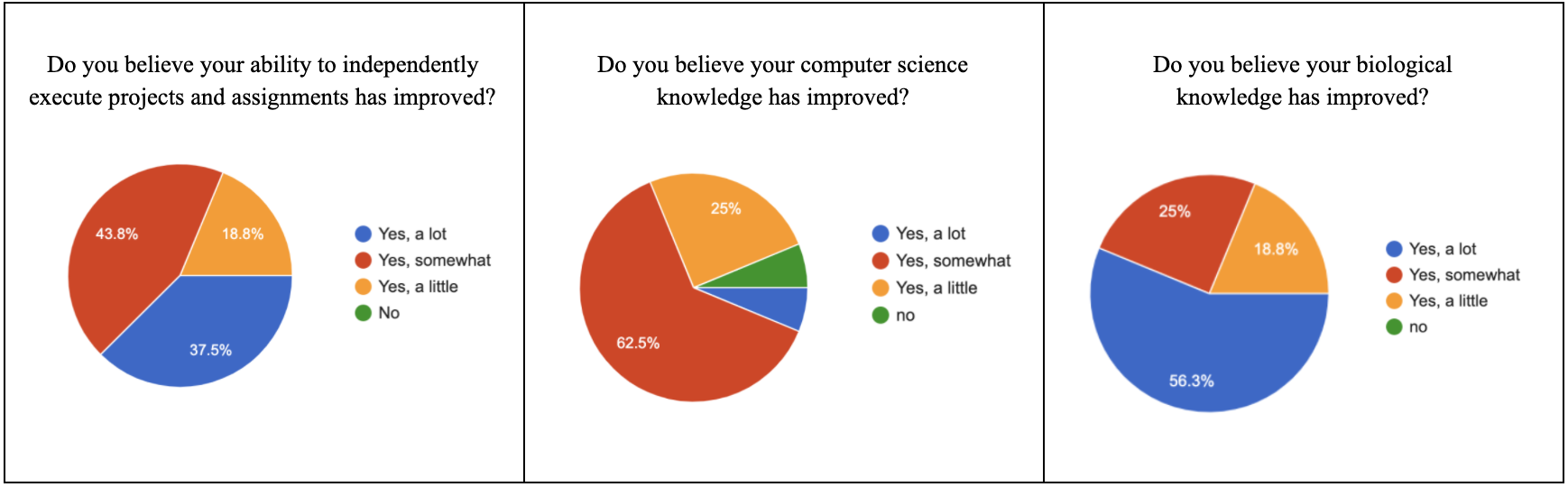
The following graphs are statistics from the program gathered in a survey sent out to participants after the program ended, proving that our online format was successful in teaching students more about computer science, biology, and scientific research.
how did the team find and select applicants?
In the outreach stage of our process, as we tried to get applicants to apply to our program, we had one primary guiding principle, and that was to make sure our outreach was widespread across the country, and not limited to only people in our network.
It would be simple for us to only reach out to local, private schools around Stanford and find a plethora of qualified applicants, but we wanted to make sure the opportunity we provided was available to a wide variety of people of all different levels, backgrounds, and locations. With an online format, there is no longer a geographical barrier, and we tried to make sure that was reflected in the outreach to potential applicants.
In May, we started to advertise our Engagement Program (all of these outreach events together) and eventually accumulated a mailing list with over 200 people on it. Then, around mid-June, we sent an email to our outreach mailing list advertising the Mentorship Program and encouraged the recipients to share widely. Before applications were due, we held an informational session over Zoom to let the applicants know what they would be applying for, and were open to feedback during the whole process to inform our ongoing design of the Mentorship Program.
In the end, we received 65 stellar applications, and accepted 20.


The questions on the application (left) looked for 4 main criteria:
1. Initiative : Because this is a relatively independent project, it was important to us that applicants showed initiative and work ethic.
2. Interest : We wanted to give this opportunity to students who showed a genuine interest in synbio and the theme of the project.
3. Feasibility : Based on students' applications and description of other commitments, we evaluated if their stated time commitment was accurate, because we wanted to be sure they could contribute meaningful time to their team's project.
4. Other Commitments : Many students lost opportunities in the pandemic, while others did not. We wanted to prioritize students who wouldn't have had another opportunity, if rejected.
how did our team ensure safety of participants?
We made sure to go through the entire process of approval for a program before the Mentorship Program began. This was done through Stanford's Protection of Minors office, where each of the mentors - 6 in total - were background checked, fingerprinted, went through a training course online detailing how to work with minors, and had to be approved by the Department of Bioengineering.
All minors involved in the program - 20 total - turned in a Liability Waiver signed by their parents. Because this wasn't an in-person program, no medical information was required. By communicating exclusively through a Slack channel which mentors could monitor, and describing clear actions mentees could take if they felt unsafe, mentors were able to ensure safe interactions between fellow participants and between participants and mentors, as outlined in the Minors Training course.

What research project did the students work on?
There is a problem in the scientific world. Many proteins that are incredibly useful in scientific research and commercial application are patent-protected, and require a licensing fee be paid to the patent-holder whenever they are used. This can be a financial burden on people trying to use the protein to do Citizen Science, or for companies attempting to use the protein for commercial applications. The project we mobilized our high school students to solve involved the following steps:
1. Identifying a patent-protected protein that would be useful if it was free-use
2. Breaking down the patents surrounding the protein
3. Compiling and aligning sequences of related proteins
4. Generating mutant sequences based on the aligned sequences
We owe the inspiration for this project to Mark Somerville and Nick Coleman at the University of Sydney. When they discovered that the famous Green Fluorescent Protein (sfGFP) is patent-protected, they embarked on a project to create a new version of GFP that had the same function, but was not protected under the patent for sfGFP. Nick and Mark gave invaluable insight regarding how to develop the workflow each team used to investigate the different patents protecting their proteins of interest, and develop free-use mutants.
how was the program structured?
Because of the remote nature of the program, the project was done mostly independently, except for meetings three times a week (Monday, Wednesday, and Friday). The meetings included the five team members and their two mentors, and were meant to coordinate work efforts and get questions answered. Therefore, willingness to take initiative and enthusiasm to learn were our top priority in applicants to our program. There was a Slack channel to connect all of the participants in the Mentorship Program, with different channels for our four teams. We also held a few workshops on the weekends to get everyone up to speed on the basic of patent law, databases, and protein biochemistry (which you can watch below).
In order to guide participants in their learning, we also gave them vague weekly "assignments" to give structure to their workflow, as they worked independently. Each of the four teams were put together based on the different levels of time commitment people in the group could afford to give, so the weekly assignment was the bare minimum individuals had to complete, with most students doing research and work outside of this. The assignments were only given in the beginning, as participants learned to use tools that were unfamiliar to them, and stopped when teams moved to focus on generating original thoughts and sequences.
Skills and Knowledge Gained From Assignments:
- How to read and interpret patents
- Basics of protein biochemistry
- Sequence alignment with clustal omega
- Using PyMol for 3D analysis of proteins
- Utilizing online databases to find patents (BLAST, NCBI)
- Navigating the protein data bank
- Python basics and use in bioinformatics problems (Rosalind)
Skills and Knowledge Gained From Team Projects:
- Working with a group of other students; team communication
- Generating original thought in order to solve a problem
- Microsoft Excel to organize workflow and information
- Coordinating and dividing work among team members
- Troubleshooting problems that came up in the process
- Presenting research findings clearly and concisely
HOW DOES THIS RELATE TO the igem team's PROJECT?
In the event that the SEED system evolves into a commercial product - and, as a startup, sells kits for viral or genetic testing based on the technology - then there could be a licensing fee to pay to the patent-holders of whatever fluorescent readout our SEED system ends up using. For example, if we decide we want to use sfGFP as our readout, then we would run into the same problem that Mark and Nick came across, and would have to pay a fee to use the protein. Currently, we are using YFP (right).
The projects we mentored high schooler students in doing can prevent this problem. We could use the same mutant-generating methods that were refined with the high-schoolers to engineer a novel version of YFP that we have currently proved to work within our system - or potentially create one with a faster diffusion rate.

While the research projects don't relate directly to the iGEM team's project (this was hard due to the remote format), both our team members and mentees learned a lot about patents, proteins, and biotechnology law throughout this process. The experience we gained in learning about patents while mentoring students throughout the summer was invaluable when we filed for a patent on our SEED technology in October, after the program had ended.
what were the results from Their projects?
Below are the final Google Slides presentations from each of the four teams, with their final results compiled. By the end of the two month project, the four teams each had between 15 and 20 novel protein sequences that shared conserved regions with their original patent-protected protein while not falling under that patent or any other one. The next steps are for us to order the DNA sequences that they have come up with, and as soon as we can we will test them out in the lab to see if they have retained the same function and can be considered novel versions of the original proteins that we can then work on moving into the public domain (a process that our wonderful mentor Drew Endy has been through before, and is willing to help us and our mentees through once we can identify the sequences in lab).
We are proud of all of our high school mentees for coming up with and executing a research project independently in less than two months! In the process, they not only generated mutant protein sequences, but they gained more knowledge in protein biochemistry and on what the research process looks like - and now if they wanted to, they could engineer novel proteins all on their own!
This year the Stanford iGEM team made a purposeful effort to reach out and start dialogues surrounding synthetic biology with a greater diversity of people. We did this in a variety of ways including a weekly synthetic biology speaker series, regular online lectures about various wet and dry and lab techniques, and a high school mentorship program that enabled 21 high school students to design free-use versions of a protein of interest. The most important part of the team's outreach, however, was the amount and diversity of people we were able to include in these conversations about synthetic biology because of our unique outreach efforts. We were able to include people ranging from university students and their families to middle school students from over 20 states and 60 cities in our live viewership alone, and even more from the published recordings of the content.

