
General Design of the Interleukin Yeast Biosensor
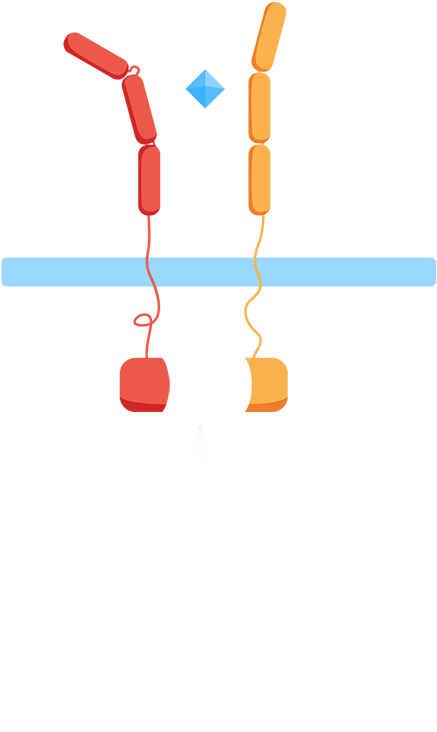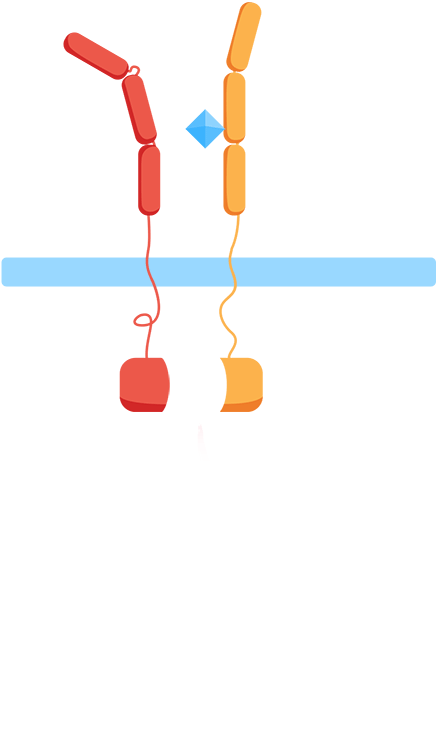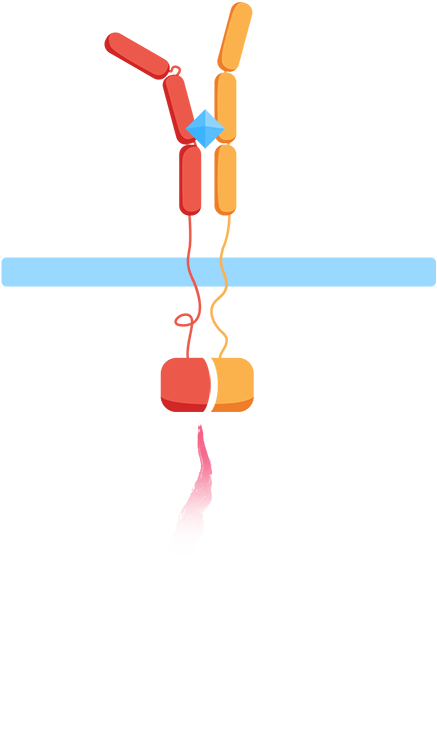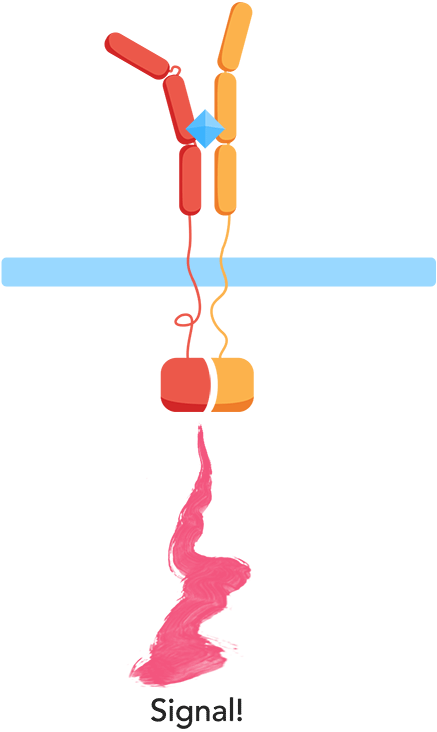
Ligand binding to the extracellular receptor domain causes a conformational change, thereby initiating gene expression of a reporter gene.
In humans, interleukin mediated signaling is implemented by the association of two or more interleukin receptor proteins, most of which fall under the category of Receptor Tyrosine Kinases (RTKs) . Association of the receptor proteins results in their autophosphorylation, and the subsequent recruitment of multiple adaptor and effector proteins that bind to the phosphorylated sites and trigger a signaling pathway .
Yeast, however, has no such receptors. .
Therefore, in order to develop a yeast-based biosensor able to monitor biomarkers for general inflammation, such as interleukins, it is essential to develop a synthetic receptor and signal transduction system in yeast. To be able to detect the vast number of different interleukins, which are all important as inflammatory markers in different contexts, we opted to establish a flexible modular platform that will enable us to easily expand our biosensor to work with different interleukins. This is in line with, and because of, the advice of the experts we reached out to in our human practices work.
As most interleukin receptors are receptor tyrosine kinases, most interleukin receptors function based on the same principle: association. Thus, we based our system on the ability of the extracellular domains of the human interleukin receptors to associate in the presence of interleukin. We have fused these domains to transmembrane helices that, when the ligand is present, come together. This physical association of the transmembrane helices brings into proximity their intracellular parts, which are fused effector domains that transmit the signal to different reporter molecules.
The general mechanism of our biosensor is summarized in the following simple steps:
Mechanism
- 1. In the presence of the biomarker of interest, the extracellular receptor domains associate.
- 2. The association of the extracellular domains results in the intracellular complementation of a split protein.
- 3. Upon complementation of the split protein, the signal is transduced and arrives at the nucleus.
- 4. As a result, a reporter gene is expressed, translating the level of interleukins into a colorimetric output.
Venturing deeper into this page will show you how we hope to achieve this by developing our very own receptor-systems!
Choice of Interleukin Receptors
To establish our system, we initially focused on three main cytokines and their receptors, IL-1α/β, IL-6 and IL-10.IL-1
IL-1
In the presence of an irritant or intruder causing inflammation, cytokines such as IL-1 are secreted by cells of the immune system in the inflamed area. When IL-1 then reaches the nearby endothelial cells, it signals to them to express certain proteins on their surface that are important for leukocyte adhesion. In this way, leukocytes in the blood are called to the site of inflammation, as they bind to the proteins the cells in this area express, before continuing their migration into the tissue . Due to the importance of this step in inflammation, and since IL-1 is so well-researched, it served as a good starting point.As mentioned above, the mechanism of action of IL-1α/β binding and signaling relies on the association of two receptors and the interleukin itself. The receptors in question are the IL-1RI and the accessory receptor IL1RAcP, where the formation of the heterotrimer between these proteins results in the activation of the pathway in the native setting .
In humans, there are two inhibitors of IL-1 signaling: the IL-1RII decoy receptor, and the IL-1 antagonist. The IL-1RII decoy receptor differs from the type 1 receptor in that it lacks an intracellular toll-like domain essential for signal relay, and, interestingly, it has a lower affinity for the IL-1 antagonist compared to the signaling IL-1RI receptor . As we only planned to use the extracellular domains of these receptors, the IL-1RII proved to be a better choice for our application, as it’d provide us with increased selectivity for the signaling IL-1 molecules, as opposed to the antagonist of the system.

IL-1 receptor binding causes the extracellular association between the IL-1 receptor accessory protein to IL-1RII.
IL-6
IL-6
Interleukin-6 is one of the most researched cytokines in inflammation, and was a natural choice for us as it stimulates the production of c-reactive protein (CRP) – a common biomarker used in clinics . In humans, the mechanism for sensing IL-6 involves the heterotrimerization of the IL-6 receptor and the co-receptor glycoprotein 130 (gp130) with the IL-6 protein. Subsequently two of the IL-6:IL-6R:gp130 trimers form a dimer. This interaction causes the intracellular parts of the two gp130 proteins to transphosphorylate each other and start a signaling cascade .We chose to try out two different designs of our extracellular IL-6 sensing modules. In one design, we link either halves of the intracellular split protein to the extracellular domains of the soluble isoform of gp130 (sgp130) via a transmembrane domain, while secreting a soluble isoform of IL-6R. In this way, the split protein will be reconstituted upon the dimerization of two heterotrimers.

Binding of IL-6 to sIL-6R leads to heterotrimerization with sgp130, with two trimers forming a dimer complex, thereby initiating the signaling cascade.
In the second design we use the association of IL-6 with one gp130 and one sIL-6R in a heterotrimer instead. Here we coupled the N-terminal half of the split protein to sIL-6R and the C-terminal half of the split protein to sgp130.

In this design, the N-terminal of the split protein is coupled to sIL-6R and the C-terminal is coupled to sgp130.
Glycoprotein 130 has six extracellular domains of which the three domains closest to the membrane mainly seem to function by positioning and bringing together the two gp130 proteins in the heterohexameric signaling complex. Since we do not depend on the association of the two gp130 proteins in the second design, we decided to use a truncated version of gp130 where we only use domain 1 to 3 in that design.
The sIL-6R has three domains; one Ig-like domain and two fibronectin-like type III domains. Only the two fibronectin-like domains (domain 2 and 3) seem to interact with the other proteins in the signaling complex . We chose to use a truncated version of the sIL-6R where we only use domain 2 and 3 since we thought this would be easier to express in our chassis.
IL-10
IL-10
Finally, we chose to look at IL-10, which is an anti-inflammatory interleukin released by macrophages after the offending agent has been removed from the site of inflammation. Another difference between IL-10 and the previous two interleukins is that IL-10 is a dimer .In IL-10 signaling, the IL-10R type I receptor associates with one domain of IL-10, and subsequently recruits the IL-10R type II. Since IL-10 is a dimer, each domain can recruit its own pair of type I and type II receptors .
Structurally, the receptor is much smaller than the previous two receptors, possibly increasing the chance of correct folding in S. cerevisiae.

Through IL-10 binding to the IL-10R type I receptor, recruitment of IL-10R type II occurs, leading to activation of the signaling pathway.
Three Biosensor Designs
Click to go to the design sections!
Common Parts of Our Biosensor Designs
As previously mentioned, the three interleukin biosensor designs share a common mechanism of action; the association of two extracellular receptors, resulting in the intracellular association of two halves of a split protein, bound to the extracellular receptors via the same transmembrane domain. Then, they implement different signal processing strategies to achieve different levels of signal processing.Transmembrane Domain
To ensure localization of the receptor protein fragments to the membrane, we used the Wsc1 endogenous yeast type 1 transmembrane domain, as this had previously been used for protein-protein interaction assays in yeast (see the ubiquitin design). For more information on how we decided on which transmembrane domain to use, see our modeling page.Transcription Factor
For two of our designs we used the synthetic transcription factor LexA-VP16 variant described by Dossani, Z. Y. et al. 2018 .This consists of the bacterial LexA DNA binding protein, fused with the viral activator domain VP16. A corresponding hybrid promoter with operator regions replaced with sequences that are recognized by LexA is also used, to avoid transcription of yeast-native genes and ensure orthogonality. The promoter we used consisted of six copies of LexO and the eno1 core promoter, in accordance with Rantasalo et. al 2016 .

Split Ubiquitin-Based Reporting
This first biosensor design builds on the split-ubiquitin based Membrane Yeast Two-Hybrid method (MYTH) , where two different proteins of interest are fused to one half of a modified ubiquitin molecule each. The modifications made to the two halves of ubiquitin render them unable to spontaneously bind to each other and reconstitute a functional protein, without first being brought together by another protein. This means that if the proteins of interest, that the ubiquitin halves are fused to, have a natural affinity for each other and consequently associate, the ubiquitin halves will also associate on the intracellular side of the membrane and reconstitute, as they’re being brought closer together by the association on the extracellular side. This suits our purposes perfectly, as we already know that the interleukin receptor domains we use have a natural affinity for each other in the presence of their corresponding interleukin. Thus, by fusing ubiquitin to each of our receptors, we know that an extracellular association will result in an intracellular reconstitution of ubiquitin.
At the same time as we fuse the two halves of ubiquitin to our two receptor
proteins, we fuse a transcription factor to the C-terminal part of ubiquitin.
As ubiquitin is recognized by deubiquitinating enzymes upon reconstitution,
this means that the deubiquitinating enzymes will cleave off the transcription
factor bound to ubiquitin, resulting in the release of the free transcription
factor into the cytosol and ultimately the cell nucleus, where it can exert
its effect.

Split ubiquitin complementation leading to release of C-ub bound LexA-VP16 to initiate gene expression.
However, as mentioned in the project description page, the concentrations
of interleukins found in sweat are rather low, and as of now, this design
has only little amplification. On the other hand, this tried and tested
method was a good, secure base to start working from.
Split TEV Protease and Membrane-bound Transcription Factor
Our intermediate design utilizes a similar receptor-system to the ubiquitin-based design. Here, our intracellular split-protein is the split TEV-protease. This is another method to monitor protein-protein interactions, described by Wehr, M. C. et al in 2006. Here, we again have two engineered inactive halves of the TEV-protease, that only regain activity when coexpressed as fusion constructs with interacting proteins . Therefore, we again utilize the receptor/TMD domains from the previous designs, but now each of our receptors will be fused to one half of the TEV-protease instead with a flexible linker.
In parallel, we also express the Wsc1 TMD, which will be sorted and localized
to the membrane. To this TMD, we’ll fuse the same transcription factor from
the previous design (LexA-VP16), and use the recognition sequence for the TEV-protease as the linker between the two.
This means that the TEV-protease, upon reconstitution, will be able to
cleave the transcription factor and free it into the cytosol. In theory,
the TEV-protease will be able to cut many transcription factors loose,
meaning that one interleukin (by extension of the association of our two
receptors) will result in the cleavage of multiple transcription factors
and thus an amplification of the signal.

Split-TEV protease complementation leading to cleavage of a TEV-recognition sequence near LexA-VP16, thereby releasing LexA-VP16 to initiate gene expression
Hijacking the Pheromone Pathway
Our most ambitious design hinges on hijacking the pheromone pathway in yeast. This design has the potential to provide a high level of amplification, which is especially important given the low concentrations of interleukins in sweat .
Yeast has a G-protein-coupled receptor (GPCR) specific to yeast mating pheromones.
When a pheromone binds to the receptor, the receptor occupancy stimulates
the G-alpha subunit of the G-protein to exchange GDP for GTP, and release
the beta and gamma subunits. The released beta and gamma subunits can
then recruit the Ste5 scaffold protein to the membrane, starting a
phosphorylation cascade eventually leading to the phosphorylation and
activation of the transcription factor Ste12 and expression of pheromone
response genes .
Since the dissociation of G-alpha from the beta and gamma subunits is what drives the pheromone cascade, we decided to design our own switch for this step in the pathway. Building onto the previous TEV protease design, we designed a mutant G-alpha with cleavage sites from the TEV protease inserted at multiple points, as described on our engineering success page. This meant that the presence of an interleukin, and ultimately the reconstitution of the TEV protease, would result in the cleavage of G-alpha into multiple fragments. These fragments would then, in theory, dissociate from the beta/gamma subunit, freeing it in a similar fashion to the normal signal transduction pathway. The free beta/gamma subunit would then be able to trigger the pheromone pathway and activate a downstream reporter gene. In order to maintain orthogonality, we used the same promoter as previously, and the transcription factor LexA-Ste12 - a synthetic transcription factor that can be activated by phosphorylation in the last step of the phosphorylation cascade in the pheromone pathway.
First, we set out to find places to insert the TEV protease cleavage sites in yeast G alpha (GPA1p). Our goals when doing this was to engineer a G alpha that
Since the dissociation of G-alpha from the beta and gamma subunits is what drives the pheromone cascade, we decided to design our own switch for this step in the pathway. Building onto the previous TEV protease design, we designed a mutant G-alpha with cleavage sites from the TEV protease inserted at multiple points, as described on our engineering success page. This meant that the presence of an interleukin, and ultimately the reconstitution of the TEV protease, would result in the cleavage of G-alpha into multiple fragments. These fragments would then, in theory, dissociate from the beta/gamma subunit, freeing it in a similar fashion to the normal signal transduction pathway. The free beta/gamma subunit would then be able to trigger the pheromone pathway and activate a downstream reporter gene. In order to maintain orthogonality, we used the same promoter as previously, and the transcription factor LexA-Ste12 - a synthetic transcription factor that can be activated by phosphorylation in the last step of the phosphorylation cascade in the pheromone pathway.
First, we set out to find places to insert the TEV protease cleavage sites in yeast G alpha (GPA1p). Our goals when doing this was to engineer a G alpha that
- 1. could bind to and exert an inhibitory function on the beta/gamma complex’s ability to trigger the signalling pathway cascade.
- 2. could, once cut by the TEV protease in the presence of a signal, dissociate from beta/gamma again, triggering the pathway.

Split-TEV protease complementation leading to cleavage of a TEV protease cut site in GPA1, thereby initiating gene expression through the yeast pheromone pathway.
Our approach can be seen in more detail under the modeling and engineering success pages.
To summarize, our last and final design entails:
To summarize, our last and final design entails:
- 1. A signaling interleukin reaches our two receptors.
- 2. The receptors associate extracellularly, giving rise to the intracellular complementation of a split TEV protease.
- 3. The now active TEV protease can cut a mutant G alpha protein into smaller peptide fragments, that’ll dissociate from the beta/gamma complex.
- 4. The beta/gamma complex can exert its recruiting function on the Ste5 scaffold protein and recruit it to the membrane, thus triggering the pheromone cascade, eventually resulting in the phosphorylation of our modified LexA-Ste12 transcription factor.
- 5. Our activated, phosphorylated transcription factor LexA-Ste12 will bind to the synthetic promoter 6xlexo eno1, and result in the transcription and translation of our reporter protein.
Why Three Designs?
This last and final design has a lot of strengths compared to the two prior designs, but also some drawbacks. Of course, changing such a conserved protein is a great challenge, as any small change could result in great consequences for its conformation and normal functions. At the same time, however, this design proved to be the best in terms of amplification andFor this reason, and to check whether our dry lab results fit with reality, all three of the abovementioned designs were tested in the wet lab, the results of which can be found on the results page.
- Zola H. Analysis of receptors for cytokines and growth factors in human disease. Dis Markers. 1996;12(4):225-240. doi:10.1155/1996/807021 ↩
- Wintheiser GA, Silberstein P. Physiology, Tyrosine Kinase Receptors. [Updated 2020 Oct 2]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2020 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK538532/ ↩
- Gunde T, Barberis A. Yeast growth selection system for detecting activity and inhibition of dimerization-dependent receptor tyrosine kinase. Biotechniques. 2005;39(4):541-549. doi:10.2144/000112011 ↩
- Kaneko N, Kurata M, Yamamoto T, Morikawa S, Masumoto J. The role of interleukin-1 in general pathology. Inflamm Regen. 2019;39:12. Published 2019 Jun 6. doi:10.1186/s41232-019-0101-5 ↩
- Fields James K., Günther Sebastian, Sundberg Eric J. Structural Basis of IL-1 Family Cytokine Signaling. Frontiers in Immunology. 2019;10:1412. doi:10.3389/fimmu.2019.01412↩
- Garlanda, C., Riva, F., Bonavita, E., Gentile, S., & Mantovani, A. (2013). Decoys and regulatory "receptors" of the il-1/toll-like receptor superfamily. Frontiers in Immunology, 4(JUL), [Article 180]. https://doi.org/10.3389/fimmu.2013.00180 ↩
- Del Giudice, Marco & Gangestad, Steven. (2018). Rethinking IL-6 and CRP: Why They Are More Than Inflammatory Biomarkers, and Why It Matters. Brain Behavior and Immunity. 70. 61-75. 10.1016/j.bbi.2018.02.013.↩
- Rose-John S, Scheller J, Elson G, Jones SA. Interleukin-6 biology is coordinated by membrane-bound and soluble receptors: role in inflammation and cancer. J Leukoc Biol. 2006 Aug;80(2):227-36. doi: 10.1189/jlb.1105674. Epub 2006 May 17. PMID: 16707558.↩
- Kurth I, Horsten U, Pflanz S, Timmermann A, Küster A, Dahmen H, Tacken I, Heinrich PC, Müller-Newen G. Importance of the membrane-proximal extracellular domains for activation of the signal transducer glycoprotein 130. J Immunol. 2000 Jan 1;164(1):273-82. doi: 10.4049/jimmunol.164.1.273. PMID: 10605021. ↩
- Zdanov A, Schalk-Hihi C, Gustchina A, Tsang M, Weatherbee J, Wlodawer A. Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon gamma. Structure. 1995 Jun 15;3(6):591-601. doi: 10.1016/s0969-2126(01)00193-9. PMID: 8590020.↩
- Mosser DM, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol Rev. 2008;226:205-218. doi:10.1111/j.1600-065X.2008.00706.x↩
- Rantasalo A, Czeizler E, Virtanen R, Rousu J, Lähdesmäki H, Penttilä M, et al. (2016) Synthetic Transcription Amplifier System for Orthogonal Control of Gene Expression in Saccharomyces cerevisiae. PLoS ONE 11(2): e0148320. https://doi.org/10.1371/journal.pone.0148320↩
- Snider, J., Kittanakom, S., Curak, J., & Stagljar, I. (2010). Split-ubiquitin based membrane yeast two-hybrid (MYTH) system: A powerful tool for identifying protein-protein interactions. Journal of Visualized Experiments. https://doi.org/10.3791/1698↩
- Wehr, M. C., Laage, R., Bolz, U., Fischer, T. M., Grünewald, S., Scheek, S., Bach, A., Nave, K. A., & Rossner, M. J. (2006). Monitoring regulated protein-protein interactions using split TEV. Nature Methods. https://doi.org/10.1038/nmeth967↩
- Hladek, Melissa & Szanton, Sarah & Cho, Young-Eun & Lai, Chen & Sacko, Caroline & Roberts, Laken & Gill, Jessica. (2017). Using sweat to measure cytokines in older adults compared to younger adults: A pilot study. Journal of Immunological Methods. 454. 10.1016/j.jim.2017.11.003. ↩
- Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26(2):339-350. doi:10.1016/j.peptides.2004.10.002↩





