Project Overview and Rationale
After interviewing the farmers, and deciding that a field-deployable genetic testing system to detect the presence or absence of the A2 allele of the β-casein (CSN2 gene), we knew we needed a detection system that could detect a single nucleotide polymorphism (SNP) that differentiates the A1 and A2 alleles of CSN2.
We researched previous testing methods and found that testing is done by sending samples to a lab, for either detection of the variant proteins in milk, or DNA sequencing (1-3). This makes this process expensive and time consuming. By developing a method to perform field testing, the time needed for testing is significantly reduced, and, if an affordable test is made, it will reduce costs. This will allow farmers to genetically identify their cows, and facilitate selective breeding for Cows with an A2 CSN2 genetic background.
To create the detection system, we identified three key steps we would have to address:
- Isolation of a DNA sample and amplification of the CSN2 alleles.
- Genotyping method to discriminate between the SNPs present in the CSN2 A1 and A2 alleles.
- Detection of an output for the genotyping method.
Our strategy is outlined in figure 1, and a summary of each step follows.
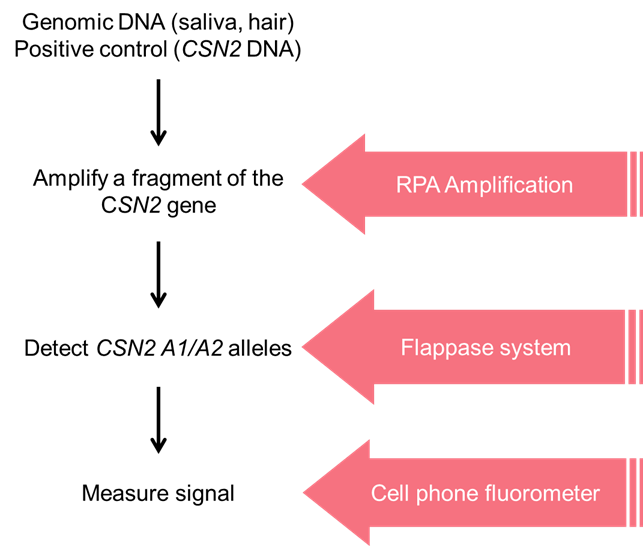
Figure 1. Summary of the strategy employed to create a field-genotyping system for the A1 and A2 alleles of the CSN2 gene.
Amplification of the CSN2 alleles
To detect the SNPs, we needed an amplification process to create an enriched sample of the CSN2 gene. We elected to use Recombinase Polymerase Amplification (RPA) (Figure 2) (4). RPA uses recombinases to form complexes with oligonucleotide primers that destabilize the DNA and facilitate invasion of the primer. Polymerase then amplifies the DNA guided by these primers. RPA differs from traditional amplification techniques such as PCR, because of its efficiency and isothermal properties. It is a fast amplification process that can be performed in as little as 40 minutes and its isothermal properties would allow the detection system to operate in the field without a thermocycler.
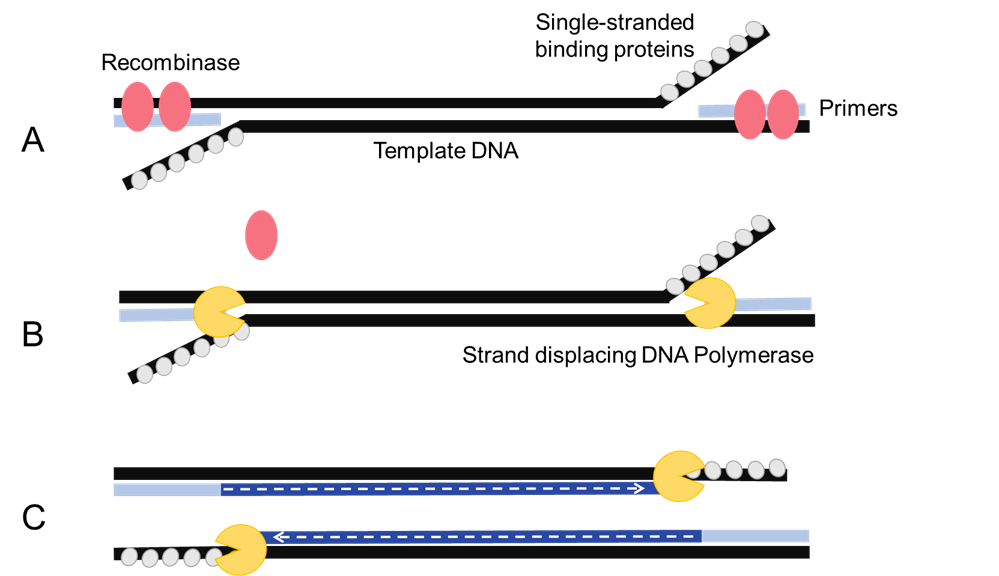
Figure 2. RPA amplification. (A) Recombinases binds to primers, and then displaces the template DNA strands for primer annealing. (B) single-stranded binding proteins stabilize the single-stranded template, and a strand-displacing DNA polymerase binds the primers and (C) synthesizes the template DNA. Multiple rounds amplify the target sequence.
For information on the progress we made on this part of the project, see the RPA Page.
Genotyping the SNPs present in the CSN2 A1 and A2 alleles
The inspiration for our genotyping system was the Invader Assay (5-7). This system uses a structure-specific 5’ flap endonucleases (Flappases) to cleave overlapping oligonucleotides at the target DNA containing the SNP (Figure 3). Some major advantages of this system include its low failure rate, the isothermal single tube reaction which can be adapted to field testing at room temperature for our farmers, and finally the level of accuracy. While the use of flap endonucleases has been validated for SNP genotyping, there is no flap endonuclease in the iGEM tool kit, inspiring use to develop the system.
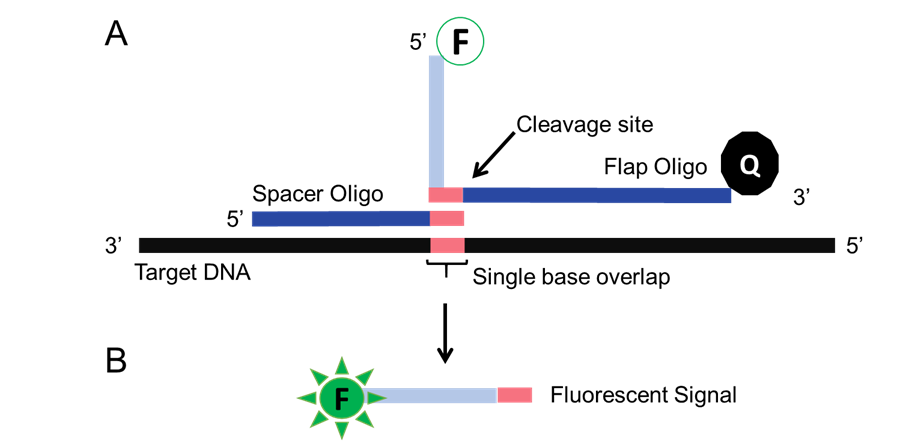
Figure 3. The Invader Assay. (A) A flap oligo, and a spacer oligo are designed that recognizes the SNP of interest. In the presence of the SNP, the oligos form a Holliday junction-like structure that creates the Flappase target site. (B) Flappase cleaves the flap oligo, releasing a fluorophore from a quencher.
For information on the progress we made on this part of the project, see the Flappase Page.
Field Implementation
In addition to having the molecular mechanism for creating a signal, we need to create field-deployable devices that farmers can use when conducting the test. Both RPA and the Invader assay will require some form of temperature control. To address this issue, we elected to create a simple heat block.
We also require a field-compatible fluorometer that can be used to detect the signal generated by the genotyping system. For this we plan on creating a cell phone based fluorometer.
For information on the progress we made on this part of the project, see the Implementation Page.
References
- A1/A2 Determination - Eurofins USA - Eurofins USA. (n.d.). Retrieved October 24, 2020, from https://www.eurofinsus.com/food-testing/services/testing-services/dairy/a1a2-casein-determination/
- Beta-casein (A2 Genotyping). (n.d.). Retrieved October 24, 2020, from https://vgl.ucdavis.edu/test/a2-genotyping
- Dairy Trait and Condition Testing. (n.d.). Retrieved October 24, 2020, from https://genomics.neogen.com/en/dairy-trait-and-condition-testing.
- Lobato, I. M., & O'sullivan, C. K. (2018). Recombinase polymerase amplification: Basics, applications and recent advances. TrAC Trends in Analytical Chemistry,98, 19-35. doi:10.1016/j.trac.2017.10.015
- Lyamichev, V., Mast, A. L., Hall, J. G., Prudent, J. R., Kaiser, M. W., Takova, T., . . . Brow, M. A. (1999). Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nature Biotechnology,17(3), 292-296. doi:10.1038/7044
- Cooksey, R. C., Holloway, B. P., Oldenburg, M. C., Listenbee, S., & Miller, C. W. (2000). Evaluation of the Invader Assay, a Linear Signal Amplification Method, for Identification of Mutations Associated with Resistance to Rifampin and Isoniazid in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy,44(5), 1296-1301. doi:10.1128/aac.44.5.1296-1301.2000
- Olivier, M. (2005). The Invader® assay for SNP genotyping. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis,573(1-2), 103-110. doi:10.1016/j.mrfmmm.2004.08.016
- Lyamichev, V., & Neri, B. (n.d.). Invader Assay for SNP Genotyping. Single Nucleotide Polymorphisms, 229-240. doi:10.1385/1-59259-327-5:229


