<!DOCTYPE html>
Contributions
Overview
When we communicated with other team members and coaches, we found that the steps of protein de novo synthesis and design are very complicated, and the shape of the domain is an important factor in protein design. In addition, there is few related website or database. Therefore, we established a domain database based on the surface shape of the domain, mainly based on the clustering and labeling of the surface shape of the domain. The contribution includes four parts: database integration, information retrieval and display, model making.
Following the spiral model, our team backtracks and updates requirements through manual operations. The model can also be used as a template for other teams to perform human practices.
Database integration
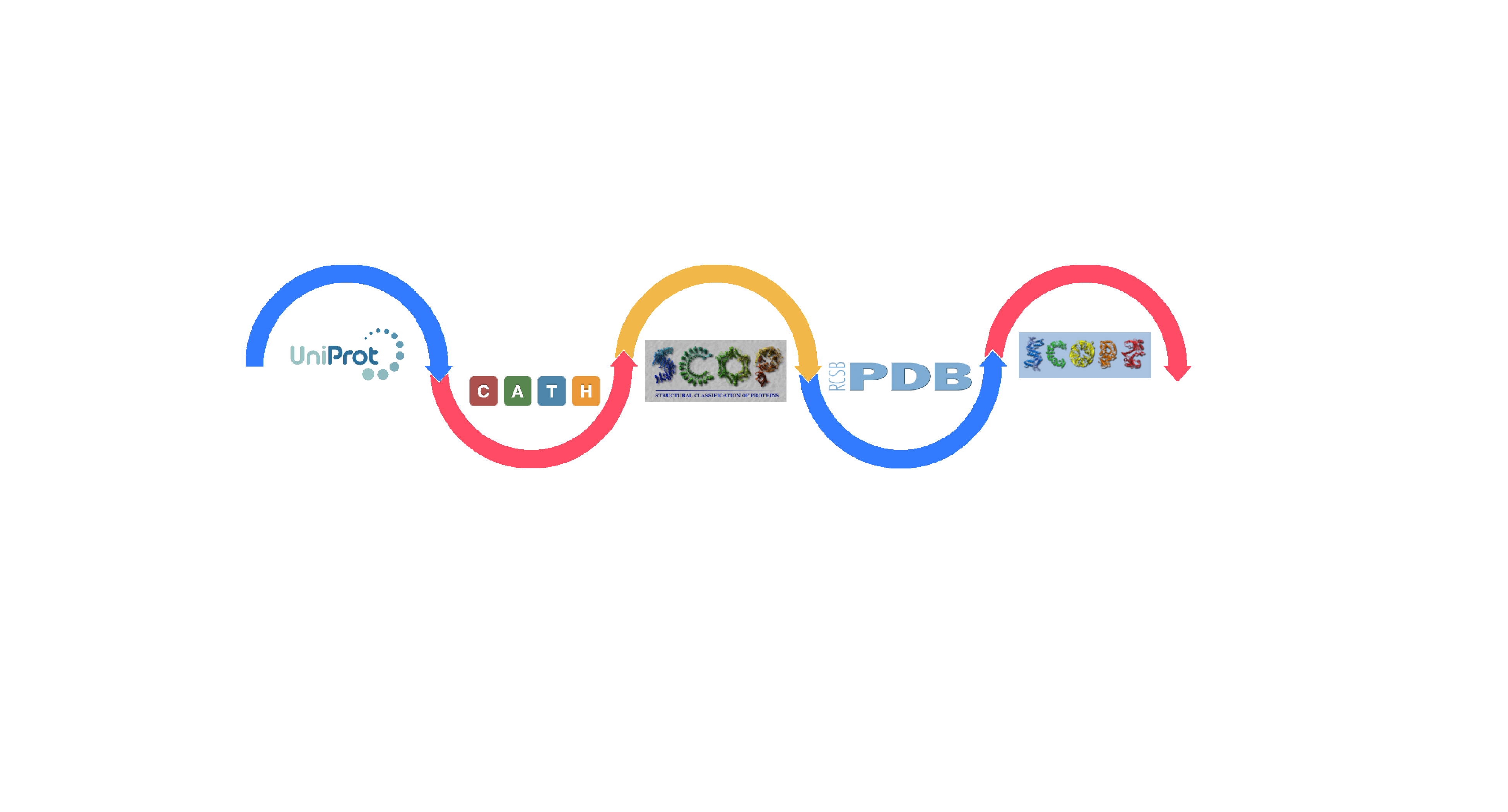
CPD3DS integrates three databases, CATH, SCOP, SCOP2 and contains many references to two databases, PDB and UniProt, as the basis for domain extraction, while providing users with as much information as possible, including sequence information, functions, etc. In addition, we provide links to other databases for each entry, such as: CATH, SCOP, so that users can quickly find the entries we provide in other databases, which can help users avoid searching for the same domain repeatedly in similar databases.
Information retrieval and display
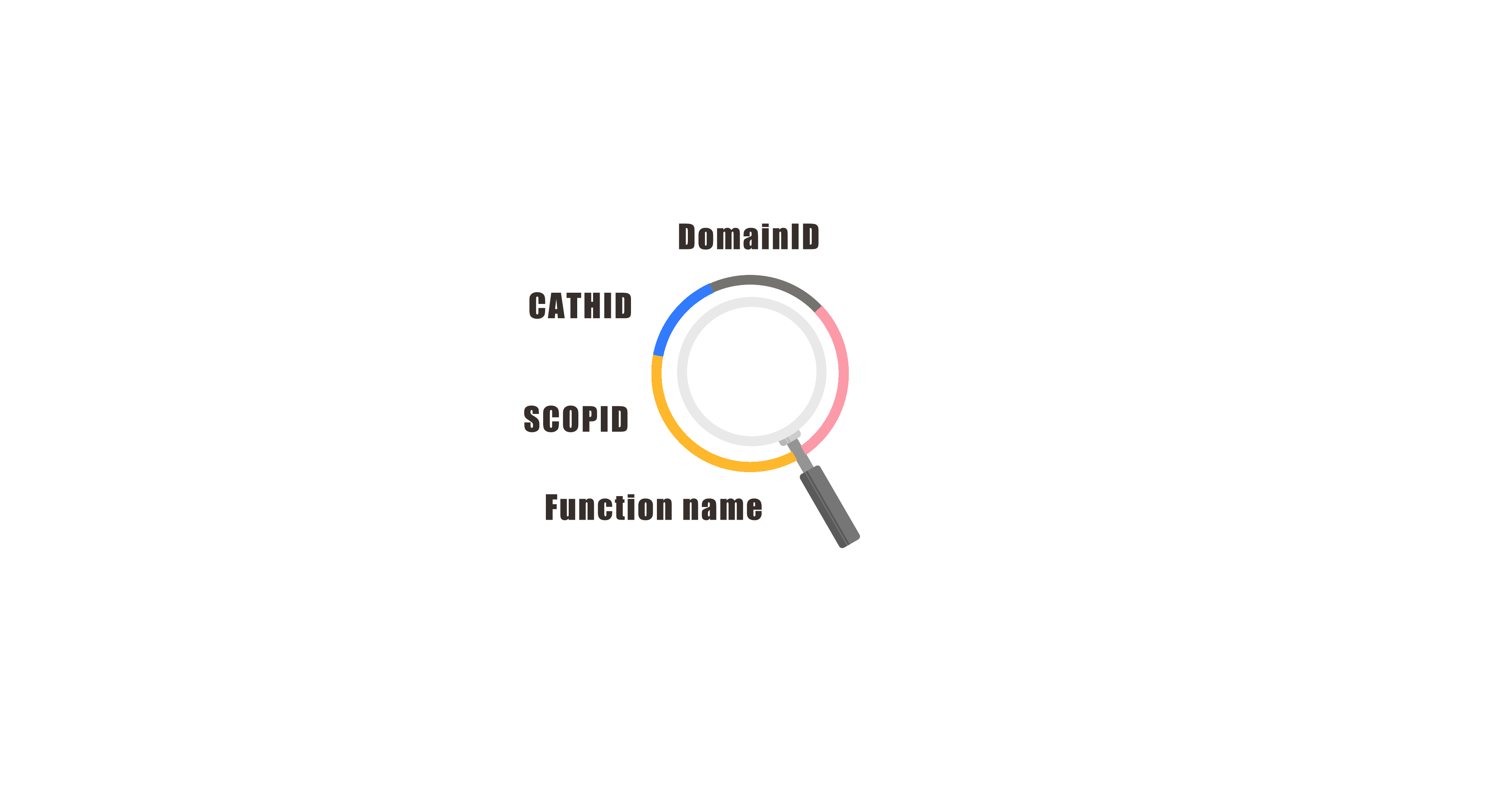
In order to accurately find useful information, CPD3DS provides three search methods. Domains can be obtained through searching by various IDs (CATHID, DomainID, SCOPID) and function name. When you forget the concrete term, you can do a fuzzy search by keywords. All the search results will be displayed directly on the web page. These work can improve the user experience significantly.
Human practice
In the process of human practice, the life cycle of software development guides our projects. We use the strategies of parallel requirements research and coding in the spiral model to promote the development of the project.
In the three-round spiral survey, meaningful suggestions were received in each round. While using suggestions to improve the project, we fully explore the scalability of the project itself. After obtaining a new version that temporarily meets the requirements, a test will be issued, and so on, until a satisfactory result is obtained.
Model making
We convert the domain into a building block model and allow users to download it on our website. Using models can not only benefit synthetic biology education, but also provide inspiration for new protein designs.


Game
In order to promote synthetic biology and iGEM to the public, we have developed the synthetic biology splicing model game protein block. In this interesting game, the key to pass the game is to use these basic knowledge to solve the corresponding problems. Players can learn some basic knowledge about synthetic biology in the process of entertainment.