Engineering Success
During our iGEM summer, our team successfully assembled many different composite parts to achieve our goal of using PRRs in a biosensing system. The main goal of these parts was to introduce the PRRs into our host organism S. cerevisiae and be able to check that expression, localization, and dimerization of our receptor/co-receptor system was functional. To achieve this, we first constructed two main types of our PRRs:
- The PRR fused to YFP, so we can check correct expression and localization using fluorescence measurements and microscopy.
- The PRR fused to the NanoLuc SmallBit, and the BAK1 co-receptor fused to NanoLuc LargeBit. These two parts are then co-expressed in S. cerevisiae to test dimerization using luminescence measurements.
Constructs with YFP
- BBa_K3610030: BAK1 with native signal peptide / YFP (BAK+)
- BBa_K3610031: BAK1 without native signal peptide / YFP (BAK-)
- BBa_K3610032: BAK1 ectodomain (eBAK)
- BBa_K3610045: EFR ectodomain (eEFR)
First, we measured fluorescence levels in S. cerevisiae samples that had been transformed with these constructs and compared them to an untransformed control group. We measured fluorescence in a plate reader assay, and also using a flow cytometer. Both experiments confirmed higher fluorescence levels in the transformed samples than in the control, indicating that the constructs are expressed correctly.

Fluorescence levels of samples with YFP constructs; Bars in yellow, green, blue and violet show expression of listed parts higher than the control.

Similar measurement done in a flow cytometer; violin charts show fluorescence intensity measured in 200000 cells per sample.
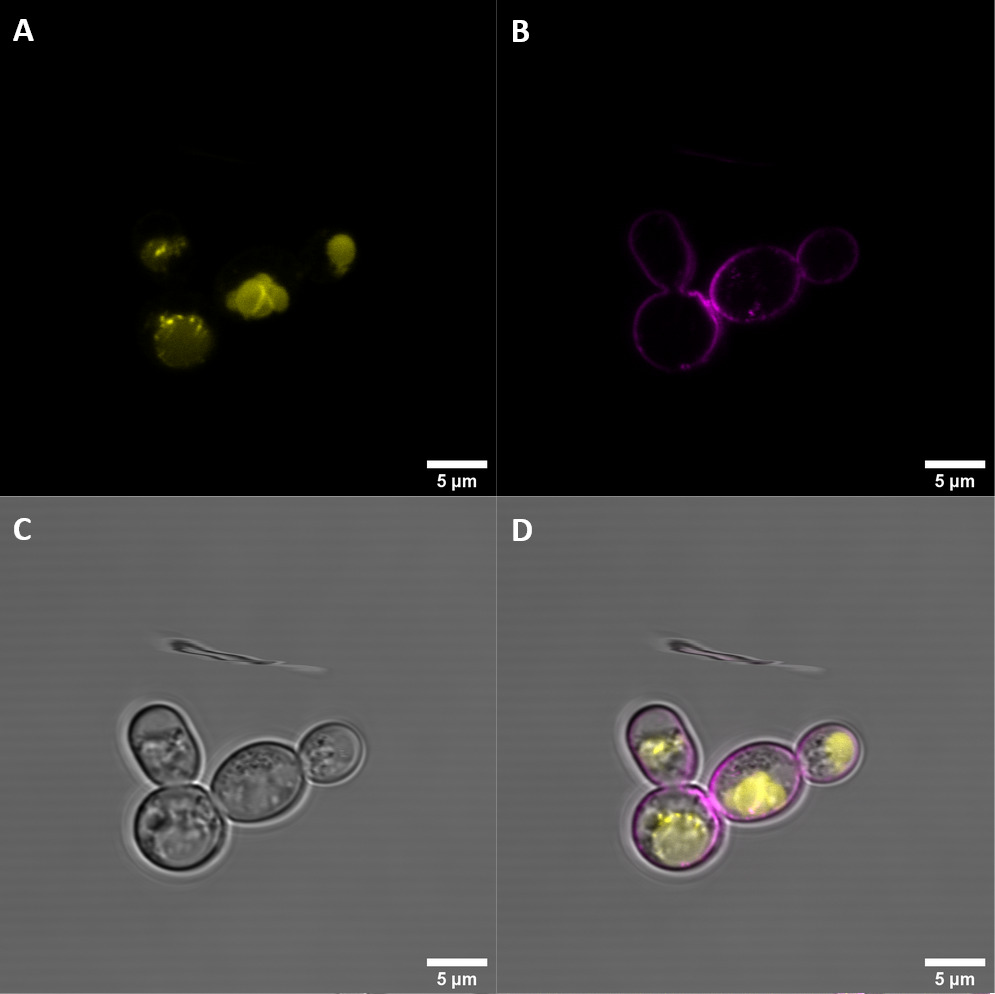
We then imaged the transformed samples with a confocal microscope to see if the constructs are correctly localized at the cell membrane. The membrane was stained with FM4-64 to make the membrane more visible. eEFR, eBAK and BAK- (which all have the yeast alpha factor signal peptide) all show correct localization at the membrane in some cells, although BAK-, which retained the kinase domain, also showed up in what seemed like the vacuoles of the yeast cells. BAK+ still has the signal peptide from A. thaliana and clearly demonstrates that this is not sufficient for correct trafficking to the cell membrane.
BBa_K3610030 (BAK+)
BAK+ shows moderate YFP fluorescence which is not co-localized with FM4-64, indicating that it is not localized at the membrane

BBa_K3610031 (BAK-)
BAK- shows strong YFP fluorescence with little co-localization with FM4-64, indicating that some gets trafficked to the membrane

BBa_K3610032 (eBAK)
Most cells show strong fluorescence, clear co-localization with FM4-64 in a ring structure in about 1% of cells

BBa_K3610045 (eEFR)
Co-localization at the membrane in about 25% of cells. Dense clumps in the cell indicate that many constructs still get stuck
From the plate reader and FACS results, we could see that our YFP constructs are expressed as expected. From the localization imaging results, we learned that the alpha-factor signal peptide is necessary to have correct localization at the cell membrane. In addition, the kinase domain also seems to play an important role in cellular trafficking, as all constructs which retained the kinase domain ended up, for the most part, inside of the cellular vacuole instead of the membrane.
Constructs with NanoLuc
- BBa_K3610038: BAK1 ectodomain fused to NanoLuc LargeBit (eEFR)
- BBa_K3610043: EFR ectodomain fused to NanoLuc SmallBit (eBAK)
After confirming good expression and localization, we assembled the composite parts with the split NanoLuc attached to test receptor/co-receptor dimerization. These two parts were then coexpressed in S. cerevisiae. With both receptor and co-receptor present at the cell membrane, addition of the EFR elicitor elf18 should drive dimerization and increase activity of the split NanoLuc system, resulting in higher luminescence. We performed multiple dimerization assays under varying conditions and test parameters.

Dimerization assay with eEFR (BBa_K3610038) and eBAK (BBa_K3610043). Shown are luminescence levels 30 min after addition of no epitope or elf18
In summary, the split parts were correctly expressed, since the transformed sample showed much higher luminescence levels than the untransformed control. We were very happy to see the NanoLuc system working in action.
However, the dimerization - and with it NanoLuc activity - did not seem to be driven by the detection of the elf18 epitope, the ligand for EFR. We are not sure why that is the case. In plants, PRRs have helper proteins that suppress dimerization when no ligand is present. The absence of these factors might explain the high activity in the absence of the elicitor in our trials.
Split NanoLuc characterization & improvement
We successfully used NanoLuc LargeBit (BBa_K1761005), and created a codon optimized version for S. cerevisiae (BBa_K3610013). These were then used in two composite parts:
- BBa_K3610054: FKBP / LargeBit NanoLuc
- BBa_K3610055: FKBP / LargeBit NanoLuc for S. cerevisiae
These parts where then each coexpressed with FRB / SmallBit NanoLuc (BBa_K3610056) to test if the codon-optimized version would result in increased luminescence levels. A dimerization assay was performed with a plate reader by adding the NanoLuc subtrate and and Rapamycin to the samples.

Luminescence levels of codon-optimized and normal NanoLuc LargeBit with and without Rapamycin
The sample using the codon-optimized version of LargeBit showed higher luminescence levels.
