Due to our inability to access lab facilities during the COVID-19 period, we decide to have a two-phase project which will be completed in 2021.
An aspect of our project which we are looking at developing further and completing is the creation of a math model. Although COVID-19 prevented us from running the necessary tests to validate our assumptions and to make progress with the creation of a concrete model, we were able to explore model designs and creation using Tinker Cell.
With tinker cell, we ran a descriptive model on our designs, and we were able to gain some more insight into the behavior of our organisms. Simultaneously, we developed a deterministic model using ordinary differential equations. However, this is still preliminary, and we are hoping to cover most of the work pertaining to the modelling in the second phase of the project when we have laboratory access.
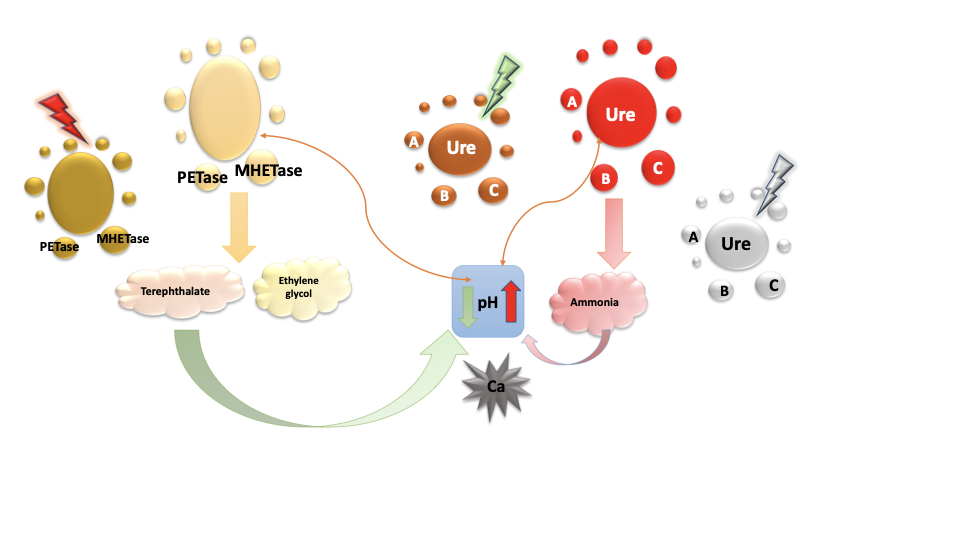
Our system is designed to be self- calibrating as shown in the image above. Hence , we set out to build a model to determine how amount of bacteria , concentration of enzymes and transcription rate affect the rate at which our system self-calibrates. To further explain the self-calibrating nature of the system: for each bacteria modified for plastic degradation, there is a bacteria modified for plastic degradation with an Alx promoter. On the other hand , for each bacteria modified for biocementation, there is a bacteria modified for biocementation with an asr promoter. This ensures that even though the products of both processes affect the ph of the system , both processes (i.e. plastic degradation and biocementation) are still able to run. Hence , the self-calibrating nature of the system
