RESULTS
Since we did not get into the lab this year, we designed our project in silico using the SnapGene software. The genetic check has results from our design in SnapGene after running an agarose simulation in SnapGene and the functional check which are predicted results from some predicted experiments to evaluate the performance of our gen constructs.
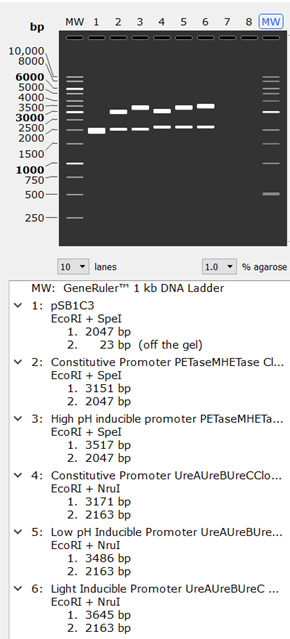
Genetic CheckFig.1
The results obtained from Agarose gel simulation in SnapGene can be found in the figure above. Using 1.0% Agarose and the GeneRulerTM 1kb DNA Ladder as the MW marker, we obtained the following bands. The first lane (1) shows the band of the vector yet to be cloned which has a total of 2070 base pairs (bp). This band is used to draw inferences when analyzing the bands of the cloned vector with inserts in the other lanes. Using the GeneRulerTM 1kb DNA Ladder, we can tell the size of each band
From the gel image, we confirmed that all inserts were properly inserted into the vector by comparing the size of the bands with that of the initial size or number of base pairs of the inserts.
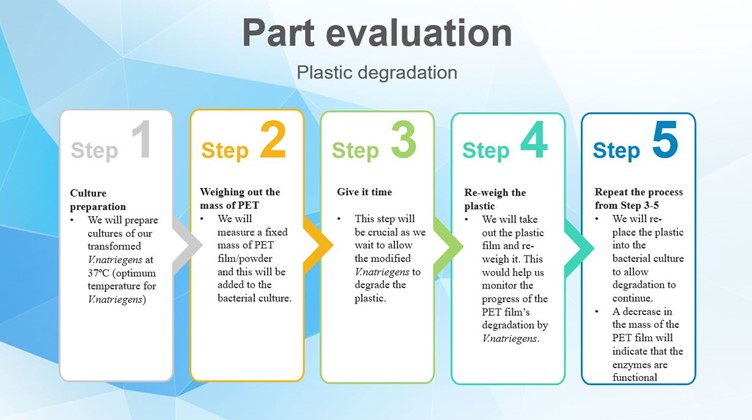
Functional CheckThe following experiments have been predicted to evaluate the processes involved in this project. Below are the experiments and graphs predicting their performance as described in the project design. The graphs however do not represent the modeling results of our project.
Fig.2: Experiment for evaluation of the plastic degradation parts.
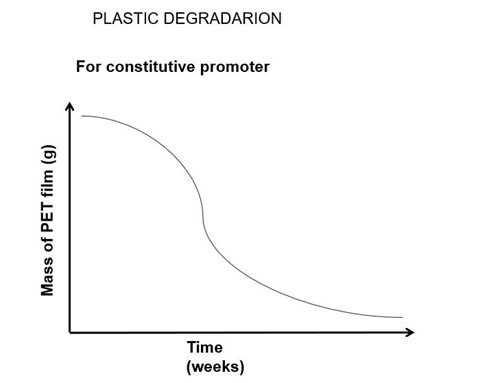
Fig.3: A graph showing the change in the mass of PET film with time.
We expect a decline in the mass of PET with an increase in time.
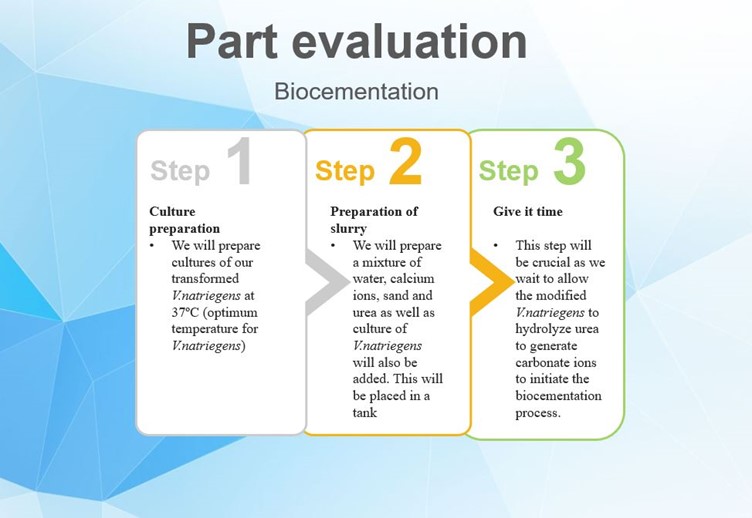
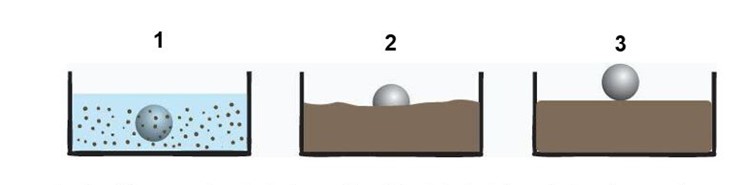
Fig.4: Experiment for evaluation of the biocementation parts
To test the mechanical strength of the bio-brick and also to monitor the progress of the biocementation process we plan to put a weighty object, for example, a metal ball into the tank at regular time intervals. As shown in the diagram we expect that the ease with which the object will sink to the bottom of the tank will decrease as time goes by.
Fig.5: A pictorial representation of the biocementation experiment.
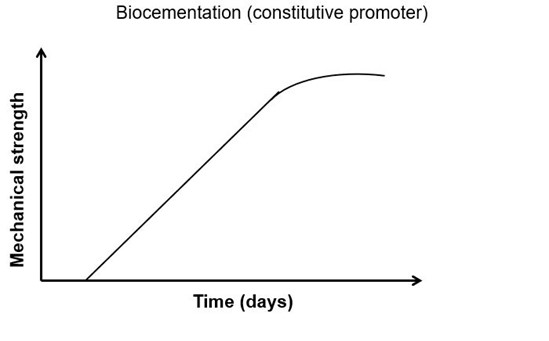
We expect a proportional relationship between the mechanical strength and the number of days. We also expect a linear increase in the mechanical strength of the slurry as time goes by until it reaches its maximum strength and plateaus.
Fig.6
For V.natriegens with pH inducible promoters (PET degradation & biocementation) and light-inducible promoters, the culture would be prepared at the pH and wavelength that will trigger the translation process for PET degradation and biocementation respectively. The experimental procedures described earlier will be applied in this situation.
Currently, we have not established an experimental technique that will aid the evaluation of the parts under an inducible promoter, but it is something we will look into.
References[1]"ISF6_4831 - Poly(ethylene terephthalate) hydrolase precursor - Ideonella sakaiensis (strain NBRC 110686 / TISTR 2288 / 201-F6) - ISF6_4831 gene & protein", Uniprot.org. Available: https://www.uniprot.org/uniprot/A0A0K8P6T7.
[2]"lipase [Ideonella sakaiensis] - Protein - NCBI", Ncbi.nlm.nih.gov. Available: https://www.ncbi.nlm.nih.gov/protein/GAP38373.1.
[3]"B.pasteurii ureA, ureB and ureC genes - Nucleotide - NCBI", Ncbi.nlm.nih.gov. Available: https://www.ncbi.nlm.nih.gov/nuccore/X78411.
[4]"ureC - Urease subunit alpha - Sporosarcina pasteurii - ureC gene & protein", Uniprot.org. Available: https://www.uniprot.org/uniprot/P41020.
[5]"ureB - Urease subunit beta - Sporosarcina pasteurii - ureB gene & protein", Uniprot.org. Available: https://www.uniprot.org/uniprot/P41021.
[6]J. Hoff, B. Daniel, D. Stukenberg, B. Thuronyi, T. Waldminghaus, and G. Fritz, "Vibrio natriegens: an ultrafast‐growing marine bacterium as emerging synthetic biology chassis", Environmental Microbiology, 2020.
[7]"Team:Harvard BioDesign/Basic Part - 2016.igem.org", 2016.igem.org, 2016. Available: https://2016.igem.org/Team:Harvard_BioDesign/Basic_Part#.
[8]"ISF6_0224 - Mono(2-hydroxyethyl) terephthalate hydrolase precursor - Ideonella sakaiensis (strain NBRC 110686 / TISTR 2288 / 201-F6) - ISF6_0224 gene & protein", Uniprot.org. Available: https://www.uniprot.org/uniprot/A0A0K8P8E7.