OVERVIEW
For developing our modified CBH-1 we utilized a hybridized catalytic domain combining the sequence from P. funiculosum and T. reesei. These modifications allow the protein to operate at more moderate conditions than the wild type. The linker sequence and cellulose-binding module were then added from the P. funiculosum wild type. These modifications were shown to increase the productivity of the protein and widen the operating parameters for the protein.
Structural models were generated to get a starting point for the protein. The predicted structure was then protonated in silico at numerous pHs. After protonation, the structures were solvated in water and underwent molecular dynamic simulation. Metrics around the simulations were taken and modelled to show protein stability within the pH 3 to 7 range.
WHAT IS CELLOBIOHYDROLASE
What impact does it have in our project
Cellobiohydrolase(CBH) is the second cellulase in our cellulose degradation efforts. After the endoglucanase has finished with the cellulose, CBH cleaves units off of the end. This is accomplished by breaking the celluloses 1,4-beta-D-glycosidic bonds. This then leaves the substrate primed for beta-glucosidase.
CBHs are commonly comprised of three different components. The first is a catalytic domain responsible for the enzymatic activity of the cellulase. Next, there is the cellulose-binding module that anchors the cellulase to the substrate. The final component is a flexible linker region that connects the two. All three of these components contribute to the efficiency, and therefore the components and their interactions with each other will be modelled for better understanding.

Figure 1. Representation of cellulase componential structure.
PRIMARY STRUCTURE
Getting the Sequence Right
For our modified CBH we looked at organisms that would best be able to provide us with a blend of high efficiency and broad operating conditions. We found the CBHs of T. reesei and P. funiculosum particularly intriguing. After discovering 'Engineering enhanced cellobiohydrolase activity,' a paper blending the two's catalytic domains and comparing the other components of CBH. The resulting domain was proven to be more effective than CBH from T. reesei and more resilient than the CBH from P. funiculosum. This hybridized catalytic domain has the following sequence.
QSAGTLQSETHPPLTWQKCSSGGTCTQQTGSVVIDANWRWTHATNSSTNCYDGNTWSSTLCPDNETCAKNCALDGAAYASTYGVTTSG
NSLSIGFVTQSNVGARLYLMASDTTYQEFTLLGNEFSFDVDVSQLPCGLNGALYFVSMDADGGVSKYPTNTAGAKYGTGYCDSQCPRDLK
FINGQANVEGWEPSSNNANTGIGGHGSCCSEMDIWEANSISEALTPHPCTTVGQEICEGDGCGGTYSDNRYGGTCDPDGCDWNPYRLGN
TSFYGPGSSFTLDTTKKLTVVTQFETSGAINRYYVQNGVTFQQPNAELGSYSGNELNDDYCTAEEAEFGGSSFSDKGGLTQFKKATSGGMV
LVMSLWDDYYANMLWLDSTYPTNETSSTPGAVRGSCSTSSGVPAQVESQSPNAKVTFSNIKFGPIGSTGNPSG
Next came the challenge of deciding on what linker and CBM to use. Luckily, this was also included in 'Engineering enhanced cellobiohydrolase activity'. We were able to see the proven improvements that accompanied these components from the P funiculosum wildtype.
STRUCTURAL PREDICTION AND INTEROGATION
What are the loops doing?
Using the Chimera and MODELLER software, we generated a starting structure file via homology modelling. This starting structure is shown below and was the cornerstone from which all of our models were built.

Figure 2.Homology structure generated for the chimeric hybridized cellobiohydrolase. Shown in rainbow.
Next came the analysis. We wanted to find out how the protein behaved with different pHs, where and when would we expect it to unravel. To simulate how the protein would react in different pHs, we changed the protonation state of the protein in silico. This was completed through the use of playmolecule.com. Playmolecule hosts a suite of computational proteomics tools and runs the software on their own servers. This is an excellent resource for iGEM teams as it does not require any hardware, and it is extremely user friendly. With the ProteinPrepare program, we gained the pdb files for our cellobiohydrolase at pHs 3, 4, 5, 6, 7, and 8. The protonated structures were then run through a nanosecond molecular dynamics simulation each. Simulations were carried out in GROMACS ver 18 on the UCalgary ARC computing cluster using gpu-v100 nodes to allow for GPU acceleration.
Molecular Dynamic Simulations were carried out by the team using the following general scheme with commands in paranthesis.
1. Convert PDB file to GRO file (gmx pdb2gmx)
2. Generate empty box to have 0.75 nanometers extra room around the protein (gmx editconf)
3. Solvate the protein with water using the spc216 water approximation (gmx solvate)
4. Generate ions to ensure the system has neutral charge (gmx genion)
5. Perform Energy Minimization (gmx mdrun with energy minimization mdp file)
6. Perform isothermal-isochoric equilibration (gmx mdrun with isothermal-isochoric equilibration mdp file)
7. Perform isothermal-isobaric Equilibration (gmx mdrun with isothermal-isobaric Equilibration mdp file)
8. Perform Molecule Dynamics (gmx mdrun with molecular dynamics mdp file)
A ten frame snapshot of the six simulations are available below.

Figure 3. Molecular dynamic simulation snapshot of modified cellobiohydrolase(green) protonated for pH 3.

Figure 4. Molecular dynamic simulation snapshot of modified cellobiohydrolase(blue) protonated for pH 4.

Figure 5. Molecular dynamic simulation snapshot of modified cellobiohydrolase(pink) protonated for pH 5.

Figure 6. Molecular dynamic simulation snapshot of modified cellobiohydrolase(yellow) protonated for pH 6.

Figure 7. Molecular dynamic simulation snapshot of modified cellobiohydrolase(salmon) protonated for pH 7.
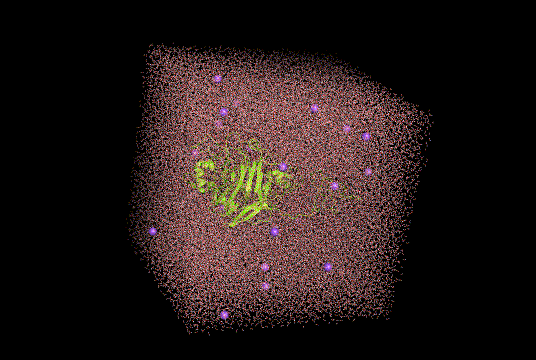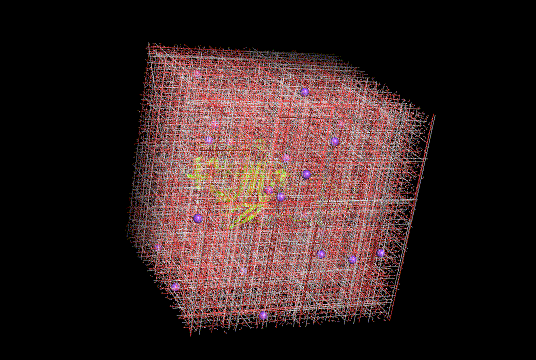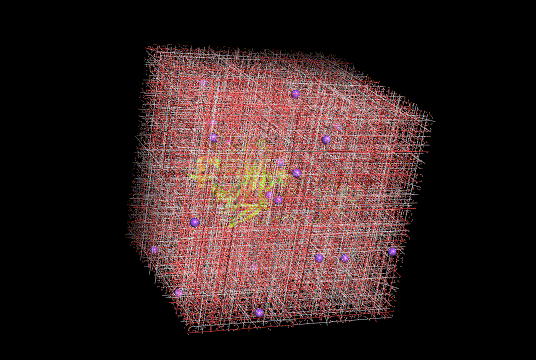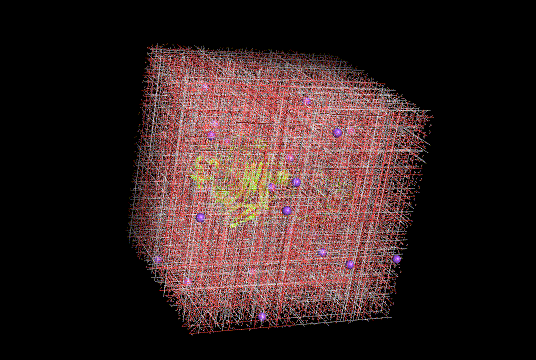
Figure 8. Molecular dynamic simulation snapshot of modified cellobiohydrolase(bright yellow) protonated for pH 8.
After the simulations, we were ready to break down the files and find metrics for comparison. Our first instinct was to make use of the GausHaus measuring software. We opted not to though in favour of RMSD. We selected RMSD for our initial metric of the dynamics as it is less computationally demanding than GausHaus. After six molecular dynamic simulations adding on anything extra may make this modelling scheme infeasible for many iGEM teams. Therefore we used the gmx rms command available within GROMACS and collected the following values.
Though the amount of data was small, we chose to conduct linear regression to correlate our change in pH and RMSD. Our first model went through all of the RMSD data points, and from the provided output, we can see that pH over the entire set was not a significant factor. However, there was still something missing. There appeared to be a slight jump in RMSD at pH 8. to test to see if this inkling was significant we ran a new linear regression with an indicator function testing for pH=8. This will allow us to see if the jump at point 8 is significant. From the output of the regression, we can see that pH 8 is a significant positive factor to the RMSD of the protein.

Figure 9. Generated R output for linear regression model. Values to note are 0.299 under Pr(>|t|) which indicates that the ordinal pH is not a significant factor in predicting RMSD.

Figure 10. Generated R output for linear regression model. Values to note are 0.035 under Pr(>|t|) which indicates that pH 9 is responsible for a significant change in predicting RMSD. This change in enumerated by 0.076 in the same column.
RESULTS
What we accomplished
From these simulations, we were able to gain insight into how the protein acts in such a broad range of pHs. We now understand how the changes in backbone RMSD is relatively constant through pHs between three and seven. Then there is a significant jump when the pH of protonation is 8. This impacts how we design and launch our system in the field. We want to set these enzymes up for success, and now we know to keep our pH south of 7.
REFERENCES
R Core Team (2019). R: A language and environment for statistical computing. R Foundation
for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Berendsen, et al. (1995) Comp. Phys. Comm. 91: 43-56.
J. Chem. Inf. Model. 2017, 57, 7, 1511–1516
Publication Date:June 8, 2017
https://doi.org/10.1021/acs.jcim.7b00190

