OVERVIEW
Yarrowia lipolytica is an up and coming yeast species gaining interest in the molecular biology community. Its oleaginous nature and its GRAS status make it a suitable chassis in food and nutrition applications. While there are synthetic biology tools for Y. lipolytica emerging in the greater scientific community, only a few iGEM teams have worked with this chassis, resulting in a limited number of Y. lipolytica parts available in the iGEM Registry of Standard Biological Parts.
While we are providing our Y. lipolytica collection to the registry, two of our parts (BBa_K3629013 and BBa_K3629016) in particular have been modelled and designed for optimal activity in Y. lipolytica. Once we have greater access to the lab, we plan to characterize these two parts (along with our other parts), and add characterization to an existing Y. lipolytica promoter in the registry (BBa_K2117000) that is important to our experimental design. Read more about our plans below!
OUR FEATURED PARTS
How these parts were made
Two of our cellulase enzyme, CBHI (cellobiohydrolase I) and EGI (endoglucanase I) were extensively modelled and optimized for function in Y. lipolytica (figure 1). This presents the first cellulases engineered for function in this yeast.
Figure 1. Homology model of Modified Trichoderma reesei EGI (BBa_K3629008) on the left and Modified Penicillium funiculosum CBHI (BBa_K3629005) on the right.
MODIFIED TrEGII
EGs function in the first step in of cellulose degradation. It works by randomly cleaving internal beta-bonds exposing new ends for further degradation by CBHs. When developing the sequence for the modified TrEGII (also known as SEGI8) (BBa_K3629016), we first focused on the catalytic domain. We utilized the SEGI-8 modified domain as it has been proven to work with higher efficiency in a broader range of conditions. For the linker, we opted to move forward with the ApCel5A linker. The ApCel5A linker was selected due to its flexibility and length, which have been shown to enhance protein efficiency with a feedstock that contains lignin (Wang et al. 2018). Finally, the wild type EGI cellulose-binding module was selected to ensure the synergy between the catalytic domain and binding module was maintained.
For the modified and wild type enzyme, we predicted the three-dimensional structure using homology modelling. From these structures, we completed molecular dynamic simulations. These simulations were then characterized by GausHaus to determine if the changes increased variance in the enzyme dynamics. The modifications actually had a net lowering of the variance in the enzyme allowing the team to move forward confident in the modifications.
MODIFIED PfCBHI
CBHs function to cleave cellobiose disaccharide units from the ends of frayed cellulose chains both from the reducing (CBHI) and non-reducing (CBHII) ends, therefore functioning as the second step of cellulose degradation. BBa_ K3629013 is a chimeric CBHI protein which we developed using a hybridized catalytic domain combining the sequence from P. funiculosum and T. reesei. These modifications allow the protein to operate at more moderate conditions for Y. lipolytica than the wild type protein. The linker sequence and cellulose-binding module were then added from the P. funiculosum wild type. These modifications were shown to increase the productivity of the protein and widen the operating parameters for the protein.
Structural models were generated to get a starting point for the protein. The predicted structure was then protonated in silico at numerous pHs. After protonation, the structures were solvated in water and underwent molecular dynamic simulation. Metrics around the simulations were taken and our modelling showed protein stability within the pH 3 to 7 range.
Future Characterization
The general characterization of the proteins will be done as per the workflow outlined in figure 2.

Figure 2. Experimental workflow of how BBa_K3629013 and BBa_K3629016 will be experimentally characterized in the future.
First the proteins will be expressed individually in Y. lipolytica. The supernatants can then be measured by ELISA to get a crude sample, or purified protein samples using the affinity tag will be made and further subjected to the Bradford Assay to determine the amount of protein purified. These samples can then by tested on their respected substrates for activity: carboxymethyl cellulose (CMC) for Modified EGI and avicel for Modified CBHI. The amount degradation of these substrates can be measured through a DNS Assay to quantify the resulting reducing sugars. Finally, we hope to compare these modified proteins to the wild-type version to verify that our mutations actually optimized function.
CHARACTERIZING BBa_K2117000
Literature-reviewed characterization
Molecular elements for Y. lipolytica such as strong constitutive promoters have not been well established in the iGEM registry. In an effort to add to the limited library of Y. lipolytica parts in the iGEM registry, we characterized the important TEF1 promoterBBa_K2117000 using the information available in the literature. TEF1 is a native promoter of the Translation Elongation Factor 1 (TEF1) gene in Y. lipolytica . TEF1 is considered to be one of the strongest native promoters found inY. lipolytica . This was confirmed when expression of the reporter gene, hrGFP, under TEF1 promoter and several other strong endogenous Y. lipolytica promoters were tested and TEF1 was shown to have one of the highest expression levels among the native promoters.
To further characterize this commonly used Y. lipolytica promoter, we gathered data on the performance of individual components of the TEF1 promoter such as the proximal promoter element (PPE), the TATA box, and the upstream activation sequence (UAS). Upon the removal of PEE, TEF1 was shown to remain functional but its strength was significantly reduced. Removal of the TATA box, however, completely diminished promoter function. Both the TEF1 TATA box and the PPE were shown to have one of the highest affinities for transcription factors in Y. lipolytica . Thus their inclusion in building hybrid promoter significantly increased promoter strength. We also found that the addition of TEF1 UAS upstream of expression construct was shown to significantly boost expression levels.
We hope that our in-depth characterization of the Y. lipolytica TEF1 promoter can make it easier for future iGEM teams to work this new exciting chassis. Using the information we provided, teams can make a more informed decision on their choice of promoter. Moreover, based on the characterization information provided on individual promoter components, teams can decide on how to play around with promoter composition to obtain their desired expression levels.
Future Characterization
We used the TEF1 promoter in a few of our genetic constructs (BBa_K3629015 and BBa_K3629018) as we needed a strong constitutive promoter to express our cellulase genes. However, we are unsure if the activity of the TEF1 promoter varies depending on the growth stage of the yeast, which would affect all recombinant protein production. Therefore, we decided to characterize the growth-phase dependent activity of the TEF1 promoter BBa_K2117000.
PART DESIGN
This part was based on the design of BBa_2117005 with the hrGFP, and was ordered from IDT with an XPR2 BBa_K3629004" terminator added at the end.
EXPERIMENTAL DESIGN
Figure 3 shows the experimental workflow that will be followed to characterize this part in the future upon further lab access.
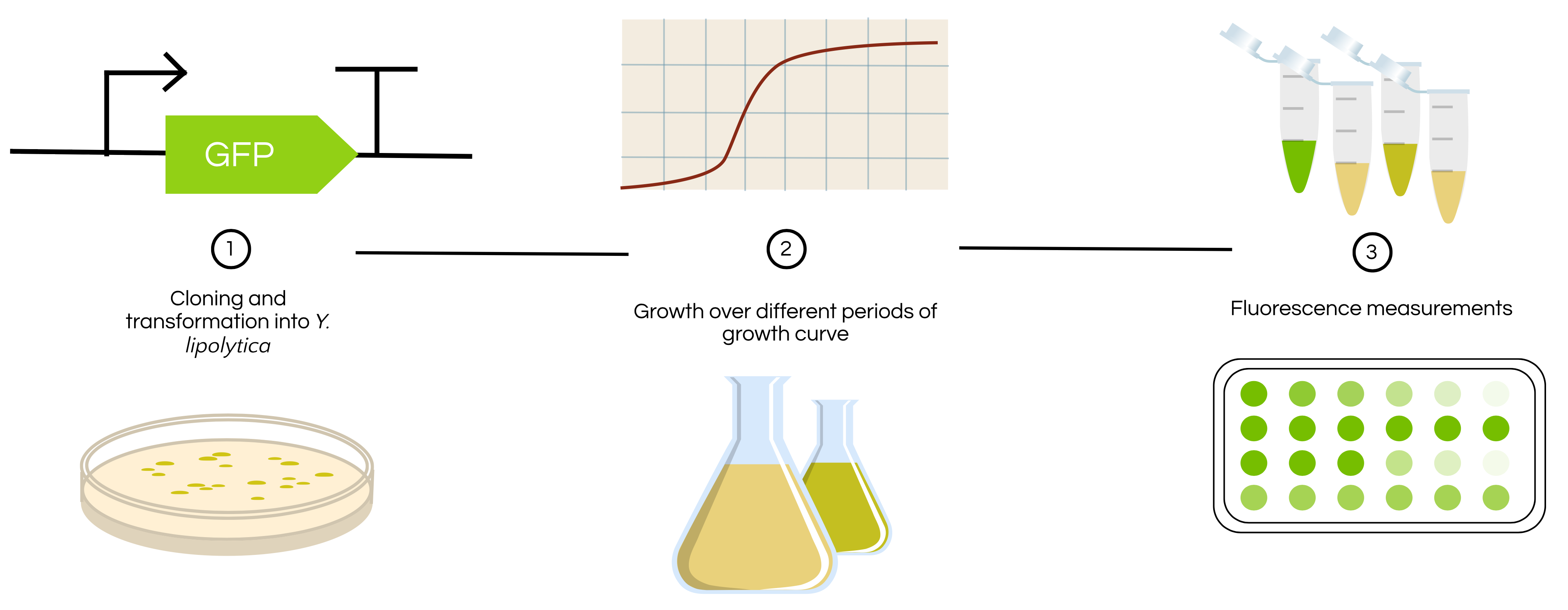
Figure 3. Experimental workflow of how BBa_K2117000 will be experimentally characterized in the future. The activity of the TEF1 promoter in Yarrowia lipolytica will be characterized based on growth state to determine how growth phase may impact promoter activity.
Protocol is as follows:
- Make a 4mL overnight culture with YPD media and a wild-type Y. lipolytica colony and grow overnight at 30ºC
- Make a 3 replicate 1:50 dilutions with YPD media with 1mL each of the overnight culture. Grow shaking at 28ºC
- Sample under fluorescence plate reader with 488 nm laser excitation and 510 nm emission, as well as OD 650, after 6, 12, 18, 24, and 30 hours
- Samples at 6, 12, and 18 hours are expected to correspond with log phase. Samples at 24 and 30 hours are expected to correspond with stationary phase.
REFERENCES
Wang, Z., Zhang, T., Long, L., & Ding, S. (2018). Altering the linker in processive GH5 endoglucanase 1 modulates lignin binding and catalytic properties. Biotechnology for Biofuels, 11(332).



