| Line 202: | Line 202: | ||
<div class = "part-design" id="part-design"> | <div class = "part-design" id="part-design"> | ||
<h2>OUR FEATURED PARTS</h2> | <h2>OUR FEATURED PARTS</h2> | ||
| − | |||
<p> Two of our cellulase enzymes: CBHI and EGI blah blah </p> | <p> Two of our cellulase enzymes: CBHI and EGI blah blah </p> | ||
| Line 234: | Line 233: | ||
</div> | </div> | ||
<p style="color:grey; font-size: 85%;"> Figure 1. Homology model of Modified <span style="font-style: italic;class="italic">Trichoderma reesei</span> EGI (BBa_K3629008) on the left and Modified <span style="font-style: italic;class="italic">Penicillium funiculosum</span> CBHI (BBa_K3629005) on the right. </p> | <p style="color:grey; font-size: 85%;"> Figure 1. Homology model of Modified <span style="font-style: italic;class="italic">Trichoderma reesei</span> EGI (BBa_K3629008) on the left and Modified <span style="font-style: italic;class="italic">Penicillium funiculosum</span> CBHI (BBa_K3629005) on the right. </p> | ||
| + | |||
| + | <h4>Modified PfCBHI</h4> | ||
| + | <p> Input stuff here </p> | ||
| + | |||
| + | <h4>Modified TrEGI</h4> | ||
| + | <p> Input stuff here </p> | ||
<br> | <br> | ||
| Line 253: | Line 258: | ||
<p style="font-size: 110%; font-weight:bold;">EXPERIMENTAL DESIGN</p> | <p style="font-size: 110%; font-weight:bold;">EXPERIMENTAL DESIGN</p> | ||
| − | <p> Figure | + | <p> Figure 2 shows the experimental workflow that will be followed to characterize this part in the future upon greater lab access </p> |
<img style="width:100%;"src="https://static.igem.org/mediawiki/2020/2/21/T--Calgary--Characterizationworkflow.png" /> | <img style="width:100%;"src="https://static.igem.org/mediawiki/2020/2/21/T--Calgary--Characterizationworkflow.png" /> | ||
| − | <p style="color:grey; font-size: 85%;"> Figure | + | <p style="color:grey; font-size: 85%;"> Figure 2. Experimental workflow of how BBa_K2117000 will be experimentally characterized in the future. The activity of the TEF1 promoter in <span style="font-style: italic;class="italic">Yarrowia lipolytica</span> will be characterized based on growth state to determine how growth phase may impact promoter activity. </p> |
<ol> | <ol> | ||
Revision as of 05:13, 26 October 2020
OVERVIEW
Yarrowia lipolytica is an up and coming yeast species gaining interest in the molecular biology community. Its oleaginous nature and its GRAS status make it a suitable chassis in food and nutrition applications. While there are synthetic biology tools for Y. lipolytica emerging in the greater scientific community, only a few iGEM teams have worked with this chassis, resulting in a limited number of Y. lipolytica parts available in the iGEM Registry of Standard Biological Parts.
While we are providing our Y. lipolytica collection to the registry, two of our parts (BBa_K3629013 and BBa_K3629016) in particular have been modelled and designed for optimal activity in Y. lipolytica. Once we have greater access to the lab, we plan to characterize these two parts (along with our other parts), and add characterization to an existing Y. lipolytica promoter in the registry (BBa_K2117000) that is important to our experimental design. Read more about our plans below!
OUR FEATURED PARTS
Two of our cellulase enzymes: CBHI and EGI blah blah
Figure 1. Homology model of Modified Trichoderma reesei EGI (BBa_K3629008) on the left and Modified Penicillium funiculosum CBHI (BBa_K3629005) on the right.
Modified PfCBHI
Input stuff here
Modified TrEGI
Input stuff here
Characterizing BBa_K2117000
Literature-reviewed characterization
Molecular elements for Y. lipolytica such as strong constitutive promoters have not been well established in the iGEM registry. In an effort to add to the limited library of Y. lipolytica parts in the iGEM registry, we characterized the important TEF1 promoterBBa_K2117000 using the information available in the literature. TEF1 is a native promoter of the Translation Elongation Factor 1 (TEF1) gene in Y. lipolytica . TEF1 is considered to be one of the strongest native promoters found inY. lipolytica . This was confirmed when expression of the reporter gene, hrGFP, under TEF1 promoter and several other strong endogenous Y. lipolytica promoters were tested and TEF1 was shown to have one of the highest expression levels among the native promoters.
Despite TEF1 showing high expression levels, a stronger promoter might be required for some engineering purposes such as metabolic engineering. Therefore, we identified methods of improving TEF1’s expression levels using tandem copies of upstream activation sequences 1B (UAS1B). Fusion of 16 copies of UAS1B to the 5’ promoter core element of the TEF1 promoter significantly enhanced the strength of the promoter. The strength of the promoter could also be further increased by fusing a truncated form of TEF1 promoter with the UAS1B copies. This is due to the fact that closer proximity of the enhancer element to the core promoter element could help improve its function.
Future Characterization
We used the TEF1 promoter in a few of our genetic constructs (BBa_K3629015 and BBa_K3629018) as we needed a strong constitutive promoter to express our cellulase genes. However, we are unsure if the activity of the TEF1 promoter varies depending on the growth stage of the yeast, which would affect all recombinant protein production. Therefore, we decided to characterize the growth-phase dependent activity of the TEF1 promoter BBa_K2117000.
PART DESIGN
This part was based on the design of BBa_2117005 with the hrGFP, and was ordered from IDT with an XRP2 terminator added at the end.
EXPERIMENTAL DESIGN
Figure 2 shows the experimental workflow that will be followed to characterize this part in the future upon greater lab access
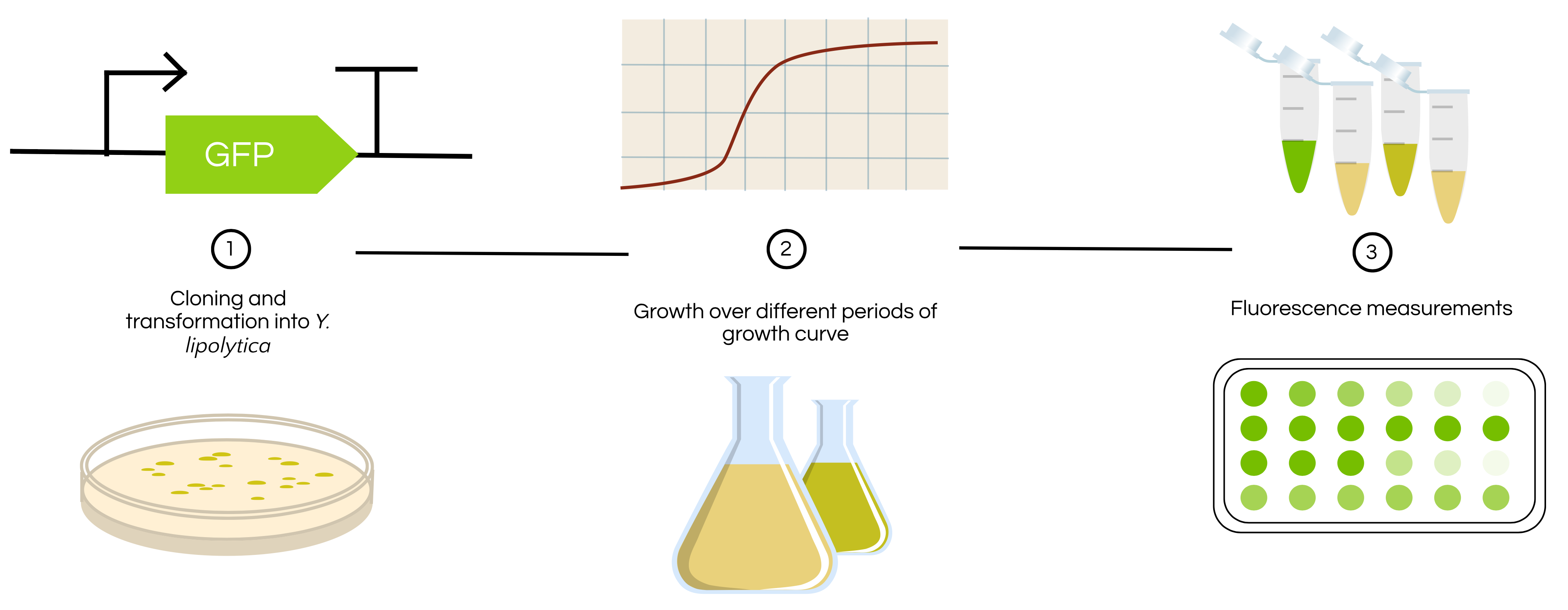
Figure 2. Experimental workflow of how BBa_K2117000 will be experimentally characterized in the future. The activity of the TEF1 promoter in Yarrowia lipolytica will be characterized based on growth state to determine how growth phase may impact promoter activity.
- Make a 4mL overnight culture with YPD media and a wild-type Y. lipolytica colony and grow overnight at 30ºC
- Make a 3 replicate 1:50 dilutions with YPD media with 1mL each of the overnight culture. Grow shaking at 28ºC
- Sample under fluorescence plate reader with 488 nm laser excitation and 510 nm emission, as well as OD 650, after 6, 12, 18, 24, and 30 hours
- Samples at 6, 12, and 18 hours are expected to correspond with log phase. Samples at 24 and 30 hours are expected to correspond with stationary phase.
REFERENCES
Next Steps
In order to provide a sustainable, community-based solution, we plan to genetically modify Rhodosporidium toruloides, an oleaginous yeast that naturally produces beta-carotene and lipids, to be more robust and resource-efficient. By modifying the yeast to produce cellulase, it can then use common agricultural waste products as an energy source for synthesizing its oil. It can then be eaten as a vitamin A supplement. The yeast strain, while naturally safe and non-pathogenic, will also be genetically modified to include a kill switch for bio-containment, and optimized for oil production.



