Background
Programming gene expression is a crucial step in studying basic human biology as well as understanding disease pathology. Our team lead Navya used CRISPR technology to mute gene expression in induced pluripotent stem cells (iPSCs). This wet lab work was done in the same research facility as 2020 Nobel Laureate Jennifer Doudna of Gladstone Institute, UCSF, who is the pioneer of CRISPR technology. CRISPR-Cas9 is a protein-RNA complex that consists of Cas9, which cuts DNA, and a small guide RNA (sgRNA) which will guide Cas9 to the desired location in the genome in the presence of a PAM sequence. Unfortunately, because of the varying efficiencies of targeting, one doesn’t know which sgRNA to use for any particular gene of interest. It is impossible to know if a sgRNA is effective until it is tested in cells. And this results in multiple sgRNAs being tested for any given application. The process of introducing this CRISPR-Cas9 complex into cells is an arduous, expensive and time consuming wet lab process. Using less efficient sgRNAs can set the research process back a month or more. With better sgRNA prediction software, this ubiquitous process pipeline can be streamlined to save time, laborious work and resources.
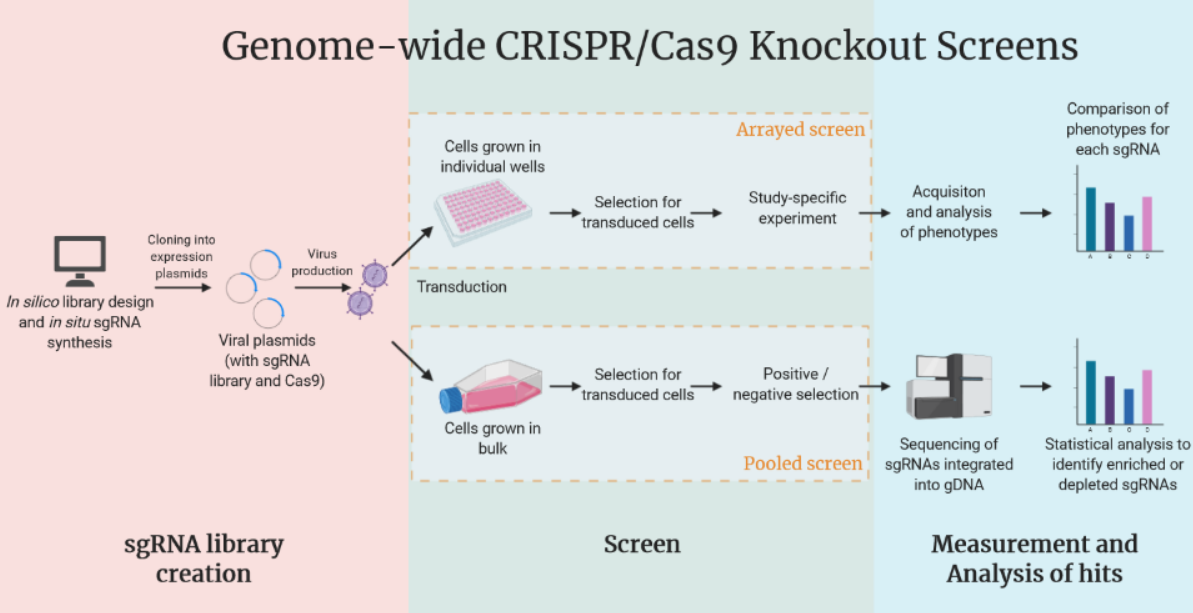

Fig-1: Genome wide CRISPR Cas9 Knockout Screen
Inspiration
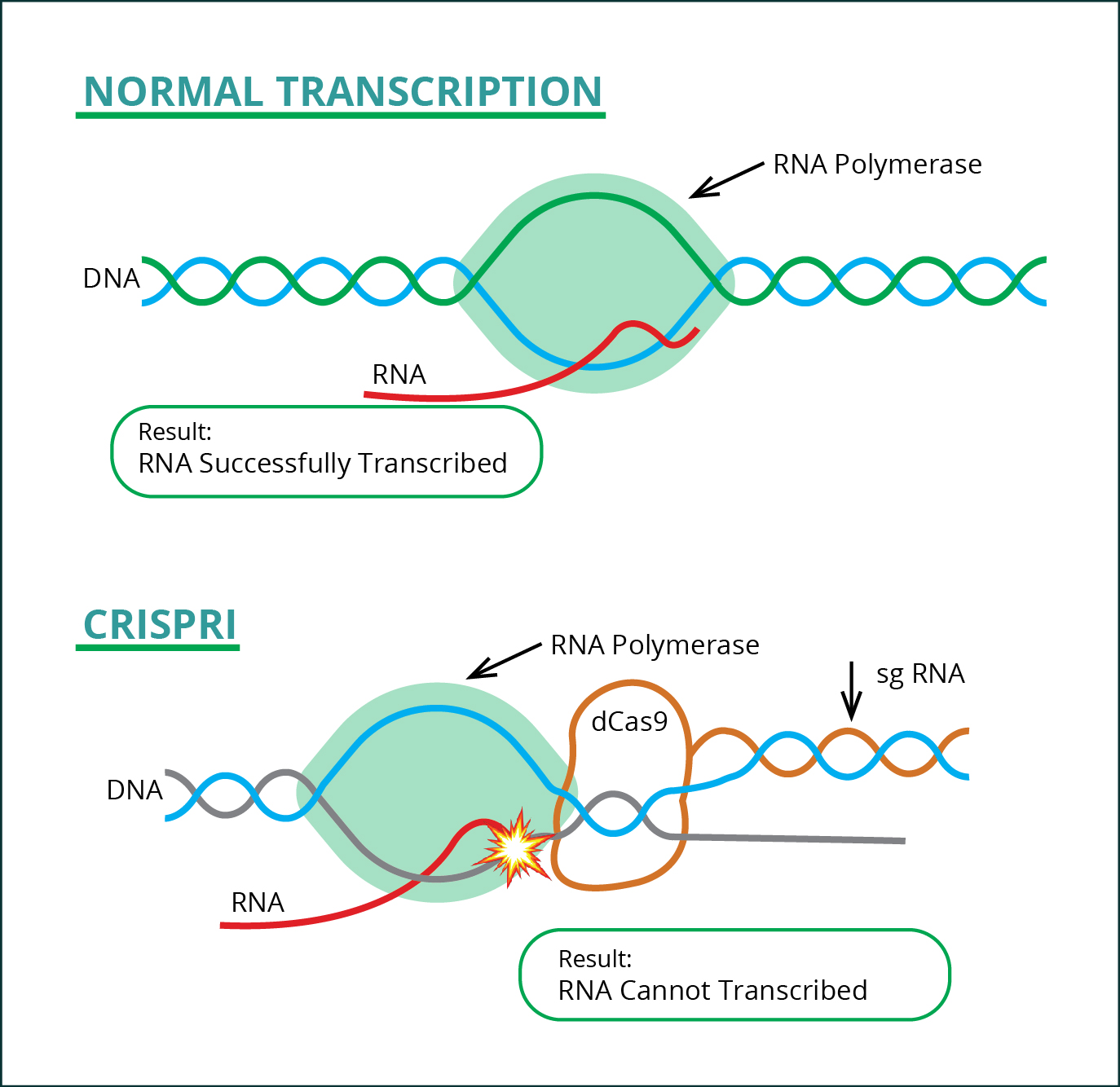
Before the project started, Dr. Perli, Navya, and the rest of the Yamanaka lab used CRISPRi (dCas9-KRAB) to study stem cell differentiation using the sgRNAs generated by Weissman’s algorithm. CRISPRi is an inactive variant of Cas9 CRISPR technology, which doesn’t have the ability to cut the DNA, but has the ability along with sgRNA to bind on target DNA in presence of PAM and it simply blocks RNA polymerase from transcribing RNA into DNA, preventing the formation of a protein. This method results in a knockdown (without altering the genome sequence) so there is no risk of incidental side effects when cutting DNA such as unwanted mutations. Furthermore, CRISPR cuts genes to render RNA ineffective, whereas CRISPRi preempts the process by preventing RNA transcription (see Figure 1). Unlike with CRISPR, simply measuring the lower amount of RNA made would demonstrate the effectiveness of the sgRNA, thereby also illuminating CRISPR’s sgRNA efficacy. We believe both CRISPRi and CRISPR share similar biophysical properties in terms of sgRNA efficacy.
 Therefore, we set out to enhance the sgRNA prediction model developed by the Weissman lab by incorporating Real-Time RNA measurements as opposed to the phenotypic data employed by the Weissman lab. This can be facilitated by using Real-Time Quantitative Reverse Transcription PCR (Real-Time qRT-PCR). Real-time PCR is a specialized technique that allows a PCR reaction to be visualized “in real time” as the reaction progresses. Quantitative PCR allows us to quantitatively measure minute sequences of DNA in a sample. So, to measure whether or RNA polymerase was prevented in CRISPRi, we use reverse transcriptase to generate the complementary DNA to the RNA whose transcription was supposed to be prevented. In this fashion, we have a reliable quantitative measurement for the success of each guide RNA (see Figure 2). Our intent for this iGEM was to improve the model in whatever ways we could, so we started by overlaying qRT-PCR data, then using machine learning to create a tool that would use wet lab sgRNA effectiveness results to tune our model. Finally, developing a computer-based algorithm during the COVID-19 Pandemic was the perfect socially-distanced research project, especially since it has been extremely difficult for a high school team to gain access to a wet-lab during this time.
Therefore, we set out to enhance the sgRNA prediction model developed by the Weissman lab by incorporating Real-Time RNA measurements as opposed to the phenotypic data employed by the Weissman lab. This can be facilitated by using Real-Time Quantitative Reverse Transcription PCR (Real-Time qRT-PCR). Real-time PCR is a specialized technique that allows a PCR reaction to be visualized “in real time” as the reaction progresses. Quantitative PCR allows us to quantitatively measure minute sequences of DNA in a sample. So, to measure whether or RNA polymerase was prevented in CRISPRi, we use reverse transcriptase to generate the complementary DNA to the RNA whose transcription was supposed to be prevented. In this fashion, we have a reliable quantitative measurement for the success of each guide RNA (see Figure 2). Our intent for this iGEM was to improve the model in whatever ways we could, so we started by overlaying qRT-PCR data, then using machine learning to create a tool that would use wet lab sgRNA effectiveness results to tune our model. Finally, developing a computer-based algorithm during the COVID-19 Pandemic was the perfect socially-distanced research project, especially since it has been extremely difficult for a high school team to gain access to a wet-lab during this time.
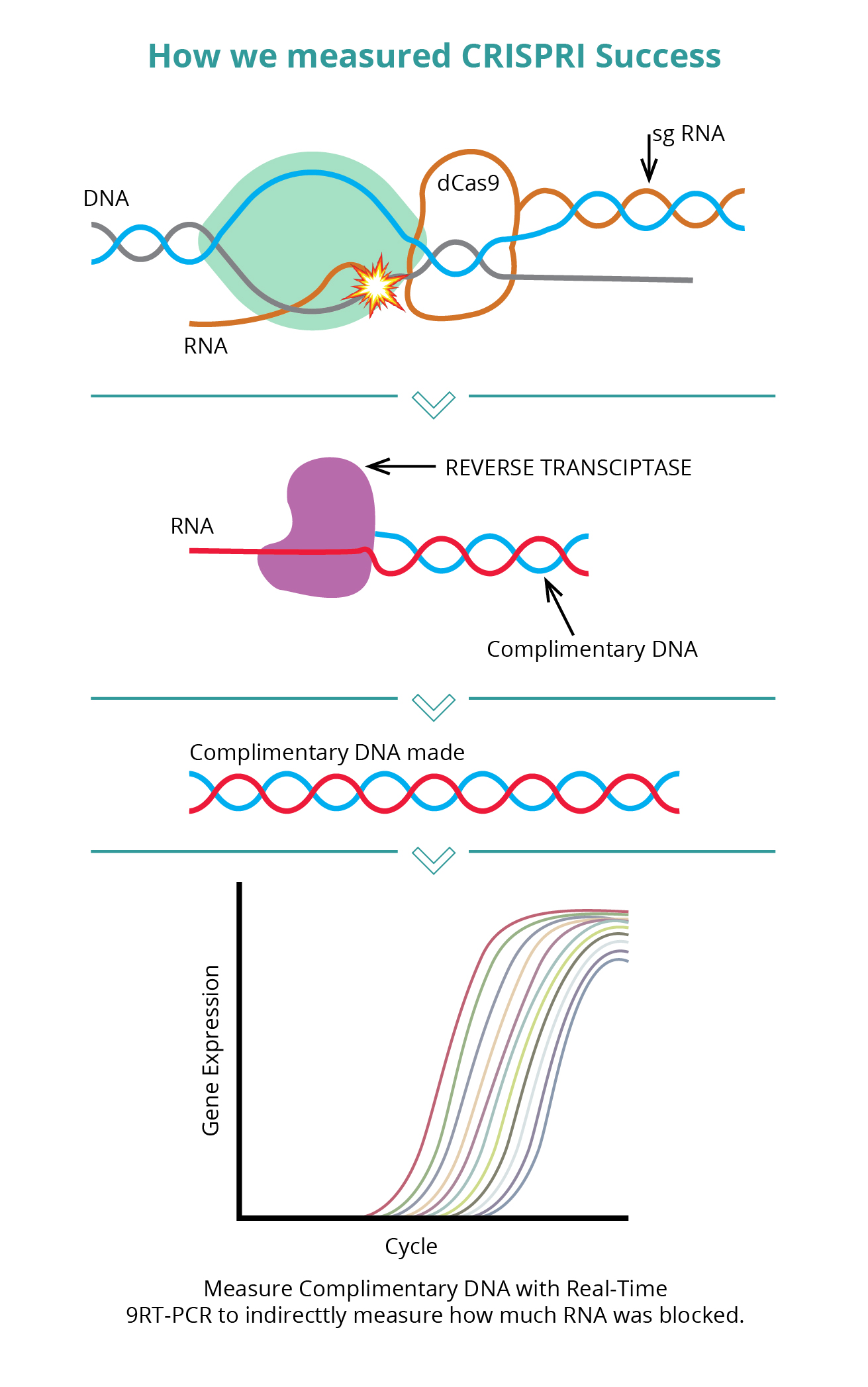

Fig-2: RNA Transcription before and after CRISPRi

Fig-3: How We Measure the Success of CRISPRi
Project
Our project worked to improve the CRIPSRi code from the Weissman lab at UCSF, which was published in 2016. Since the program was initially written in Python 2.7, the code was too outdated for new updates to be made so we changed the code to Python 3.8. Our updated version of the Weissman code improved on existing algorithms to predict sgRNA accuracy by machine learning models to select the optimal guide RNA. These models used quantitative measurements as a promising option and feature new methods to improve sgRNA efficacy. The model overlaid the qRT-PCR data with the current code to avoid overfitting the data. Our project was lacking in amount of Real-Time qRT-PCRdata (we were only able to analyze 18 genes each with 10 sgRNAs) data because, as previously mentioned, the implementation of CRISPRi is a lengthy process, which implies that obtaining data is also a lengthy process. We hope in the future to find a user-friendly method of receiving reliable data on the effectiveness of sgRNAs to further enhance our machine learning models. Another outdated aspect of the code was its dependency on an older version of the human genome data, hg19, and so, we found the updated version that was compatible with our code, hg38. Furthermore, our team collaborated with other labs to predict sgRNA efficacy for additional genomes, such as the Bovine genome. Lastly, we increased the accuracy of the Weissman CRISPRi library by incorporating new variables: the square of the distance from the transcription start site. We also had greater off-target stringency by analyzing the whole genome for off-targets, including the mitochondria.
Future Work / Improve
We are working on developing a web server for easy input that could be developed and integrated into our system. Additionally, the updated code has increased the potential of the algorithm to work in other applications, so the code can be implemented easily into future ideas and projects. The algorithm can be built upon and its efficiency improved by including data from testing with tissue-specific sgRNAs and/or host-specific sgRNAs as well. Other dependent parameters can be further tuned with more experiments. From a software perspective, the software itself can be enhanced by integrating deep learning algorithms like RNN which could potentially make the model’s prediction better. Scoring mechanisms like Azimuth could also be incorporated to improve the scores of the guide RNAs. And, as we mentioned earlier, more Real Time qRT-PCR data can be added to improve the model. The model will already take qRT-PCR data (granted, it needs to be the same format which we used) and use it to enhance the model. In the future, when reputable labs input Real-Time qRT-PCR data from knockdowns into our models, we can have a worldwide collaborative effort to improve the sgRNA algorithm slowly but surely!
Works Cited
Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, Weissman JS. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife. 2016 Sep 23;5:e19760.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816-21.>
Nolan, Tania, et al. “Quantification of MRNA Using Real-Time RT-PCR”. Nature News, Nature Publishing Group, 9 Nov. 2006, www.nature.com/articles/nprot.2006.236.
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013 Feb 28;152(5):1173-83.
Wong, Alan S. L., et al. “Multiplexed Barcoded CRISPR-Cas9 Screening Enabled by CombiGEM”. Proceedings of the National Academy of Sciences, vol. 113, no. 9, 10 Feb. 2016, pp. 2544–2549, 35047b0b-d7cb-4707-9c5f-4a4bb36b6286.filesusr.com/ugd/99161c_be25eeceea784d11972e5da3b4caa94f.pdf, 10.1073/pnas.1517883113. Accessed 17 July 2020.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816-21.>
Nolan, Tania, et al. “Quantification of MRNA Using Real-Time RT-PCR”. Nature News, Nature Publishing Group, 9 Nov. 2006, www.nature.com/articles/nprot.2006.236.
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013 Feb 28;152(5):1173-83.
Wong, Alan S. L., et al. “Multiplexed Barcoded CRISPR-Cas9 Screening Enabled by CombiGEM”. Proceedings of the National Academy of Sciences, vol. 113, no. 9, 10 Feb. 2016, pp. 2544–2549, 35047b0b-d7cb-4707-9c5f-4a4bb36b6286.filesusr.com/ugd/99161c_be25eeceea784d11972e5da3b4caa94f.pdf, 10.1073/pnas.1517883113. Accessed 17 July 2020.

