Quality of guide RNAs can be measured only after doing a lab experiment. But when you cannot get to a lab, we can use software to do quality scoring. While our goal is to design and build a software tool based on machine learning, we wanted to start based on something that already exists.
After research, we found a machine learning based algorithm used by WeissMan’s lab.
(https://github.com/mhorlbeck/CRISPRiaDesign)
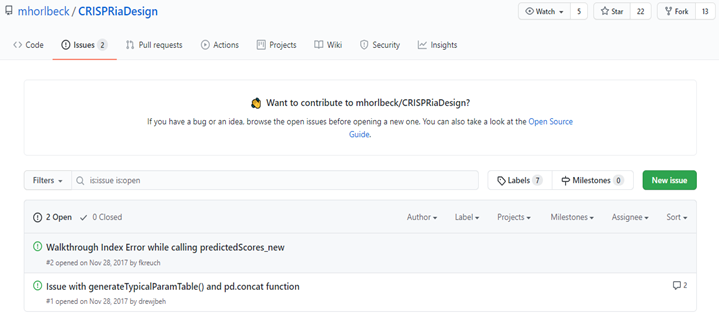
But that source code is very old and currently not functional. It can be seen from below snippet that there are others who also want to use same tool but having issues that are not fixed in years.
It is a very interesting and exciting experience on how we finally could take it to a level that can be a base for our project.
After research, we found a machine learning based algorithm used by WeissMan’s lab.
(https://github.com/mhorlbeck/CRISPRiaDesign)
But that source code is very old and currently not functional. It can be seen from below snippet that there are others who also want to use same tool but having issues that are not fixed in years.

It is a very interesting and exciting experience on how we finally could take it to a level that can be a base for our project.
Phase 1
Researched into different modules used by the above project and tried to understand what those modules are, what kind of algorithms are used.
We Designed AWS Based Cloud Platform that is accessible to the entire team over the internet and setup an environment as mentioned in the project.
We started the Development on Python 2.7 as mentioned in the project and fixed a few initial bugs we encountered.
When we Tested The flow stopped at creating Secondary structures relating to RNA.
We Designed AWS Based Cloud Platform that is accessible to the entire team over the internet and setup an environment as mentioned in the project.
We started the Development on Python 2.7 as mentioned in the project and fixed a few initial bugs we encountered.
When we Tested The flow stopped at creating Secondary structures relating to RNA.

Phase 2
We Researched into ViennaRNA again and finally found issues with Python version incompatibility.
We Designed again a virtual environment with Python version 3.8 in the same cloud environment.
Now we ended up with a Development effort of porting all current modules of above github project into 3.8.
When we started Testing the flow stopped now at a different stage where Off-target stringent related code required few bowtie index files to test alignment.
We Designed again a virtual environment with Python version 3.8 in the same cloud environment.
Now we ended up with a Development effort of porting all current modules of above github project into 3.8.
When we started Testing the flow stopped now at a different stage where Off-target stringent related code required few bowtie index files to test alignment.

Phase 3
We had to Research into bowtie indexes that need to be built for different flank sequence regions and other regions.
We had to take help our Dr. Perli to understand the semantics of these indices and Design our environment again with appropriate bowtie modules.
We Created bowtie indices files by downloading appropriate genomic data and then made code modifications as part of Development to use these indices.
When we Tested we finally landed with some code that can be base for our main project which tuning algorithm after training with Quality Data.
In these different phases we Learnt
In this whole process we Improved the current code and brought it to a stage that can be used by others. Our project based on this base and works speaks itself of this Engineering Success!!!
We had to take help our Dr. Perli to understand the semantics of these indices and Design our environment again with appropriate bowtie modules.
We Created bowtie indices files by downloading appropriate genomic data and then made code modifications as part of Development to use these indices.
When we Tested we finally landed with some code that can be base for our main project which tuning algorithm after training with Quality Data.
In these different phases we Learnt
- How to setup and work in AWS Cloud Environment
- Machine algorithms like lasso/Ridge/ElasticNetCV
- Statistical Analysis of Data
- What is Vienna RNA
- Various versions of bowtie indexes how they help with faster alignment testing instead of going through the entire genome.
In this whole process we Improved the current code and brought it to a stage that can be used by others. Our project based on this base and works speaks itself of this Engineering Success!!!


