CRISPRi is a powerful tool for modulating gene expression in human cells. By designing a gRNA homologous to the target gene of interest, one can achieve targeted knockdown of the specific gene of interest. However, with current methodologies, one has to screen multiple gRNA sequences for efficient targeting while minimizing off-target effects.
Our software presents a prediction model for identifying the best gRNA sequence for efficient gene targeting in human cells. Starting with experimental data from knocking down specific genes using several gRNAs in iPS cells, we leverage machine learning to inform better selection of the gRNA.
This software tool will be invaluable for designing gene targeting gRNAs and will reveal underlying biochemical principles governing CRISPR efficiency.
It is an improvement to works done by Weissman’s lab (Horlbeck et al., eLife 2016) and available at https://github.com/mhorlbeck/CRISPRiaDesign
Our Software has multiple components that are represented as notebooks that can be run in any jupyter notebook server.
It can be installed and set up easily by following instructions mentioned in the README where our software is available with open source license.
The release can be download from the following url https://github.com/igemsoftware2020/GunnVistaPingry_US/releases/tag/1.0.0
Our software presents a prediction model for identifying the best gRNA sequence for efficient gene targeting in human cells. Starting with experimental data from knocking down specific genes using several gRNAs in iPS cells, we leverage machine learning to inform better selection of the gRNA.
This software tool will be invaluable for designing gene targeting gRNAs and will reveal underlying biochemical principles governing CRISPR efficiency.
It is an improvement to works done by Weissman’s lab (Horlbeck et al., eLife 2016) and available at https://github.com/mhorlbeck/CRISPRiaDesign
Our Software has multiple components that are represented as notebooks that can be run in any jupyter notebook server.
It can be installed and set up easily by following instructions mentioned in the README where our software is available with open source license.
The release can be download from the following url https://github.com/igemsoftware2020/GunnVistaPingry_US/releases/tag/1.0.0
Odigos Guide Predictor
One just needs to specify the GENES_LIST_TO_FIND_GUIDES option in iGEM_CRISPRi_Gene_Guide_Selector.ipynb notebook. Then select kernel->Restart & Run All in notebook server. It gives the top 10 guides for each of genes after considering off-target stringency.
Ex: GENES_LIST_TO_FIND_GUIDES = ['APOL1','APOL3','PRRC2A']
Snippet from notebook:
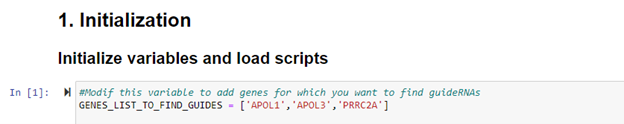
Then it runs based on the predictor already trained and generated by Odigos Model Generator.
A partial snippet of above query result will be somewhat like below.
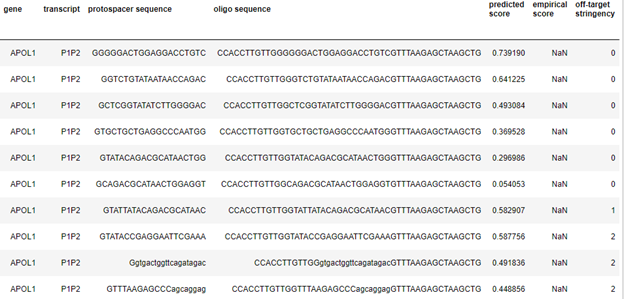
Ex: GENES_LIST_TO_FIND_GUIDES = ['APOL1','APOL3','PRRC2A']
Snippet from notebook:

Then it runs based on the predictor already trained and generated by Odigos Model Generator.
A partial snippet of above query result will be somewhat like below.

Odigos Model Generator
This tool can be used to train and generate new models. This component along with underlying software components are well documented so that it can be further extended, modified to create different ML and AI Based tools.
Odigos Model Compare
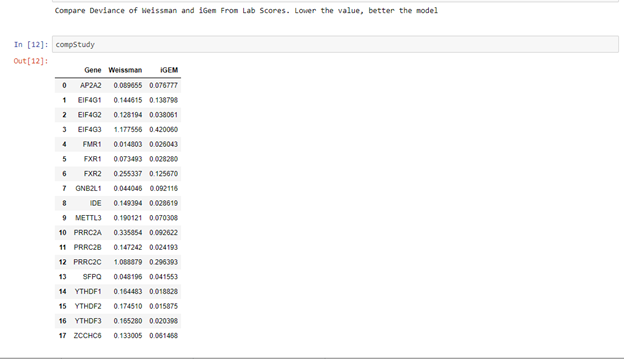
This tool can be used to do a comparison study of the guideRNA activity scores predicted by Weissman’s algorithm, our algorithm and lab scores.
All that needs to be done is to just modify one option in the notebook iGEM_CRISPRi_sgRna_Score_Comparision.ipynb and simply run Kernal->Restart & Run all from Jupyter Notebook.
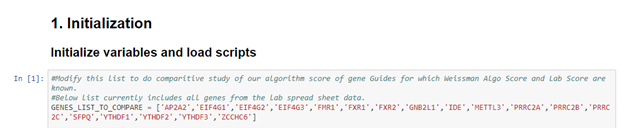
Then results will be displayed somewhat like below.
Works Cited Horlbeck, Max A, et al. “Compact and Highly Active Next-Generation Libraries for CRISPR-Mediated Gene Repression and Activation". ELife, vol. 5, 23 Sept. 2016, p. e19760, elifesciences.org/content/5/e19760, 10.7554/eLife.19760. Accessed 28 Oct. 2020.
All that needs to be done is to just modify one option in the notebook iGEM_CRISPRi_sgRna_Score_Comparision.ipynb and simply run Kernal->Restart & Run all from Jupyter Notebook.

Then results will be displayed somewhat like below.

Works Cited Horlbeck, Max A, et al. “Compact and Highly Active Next-Generation Libraries for CRISPR-Mediated Gene Repression and Activation". ELife, vol. 5, 23 Sept. 2016, p. e19760, elifesciences.org/content/5/e19760, 10.7554/eLife.19760. Accessed 28 Oct. 2020.

