Introduction
Weissman’s algorithm model is trained based on phenotype based screening experiment data. Odigos iGEM algorithm is trained using qRT-PCR Data and different feature parameters are tuned. There are 3 Key Modules in this application.
- Odigos Model Generator
- Odigos Model Compare
- Odigos Guide Predictor

Odigos Model Generator
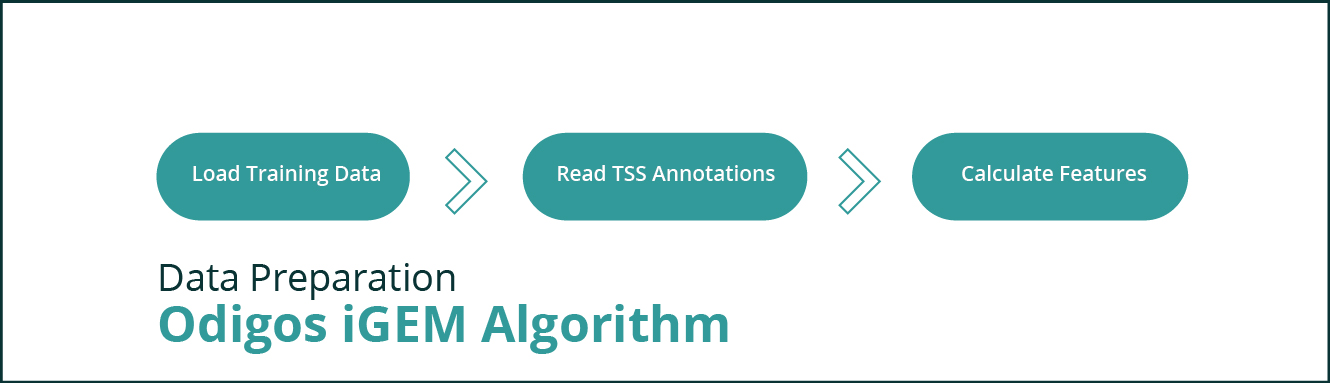
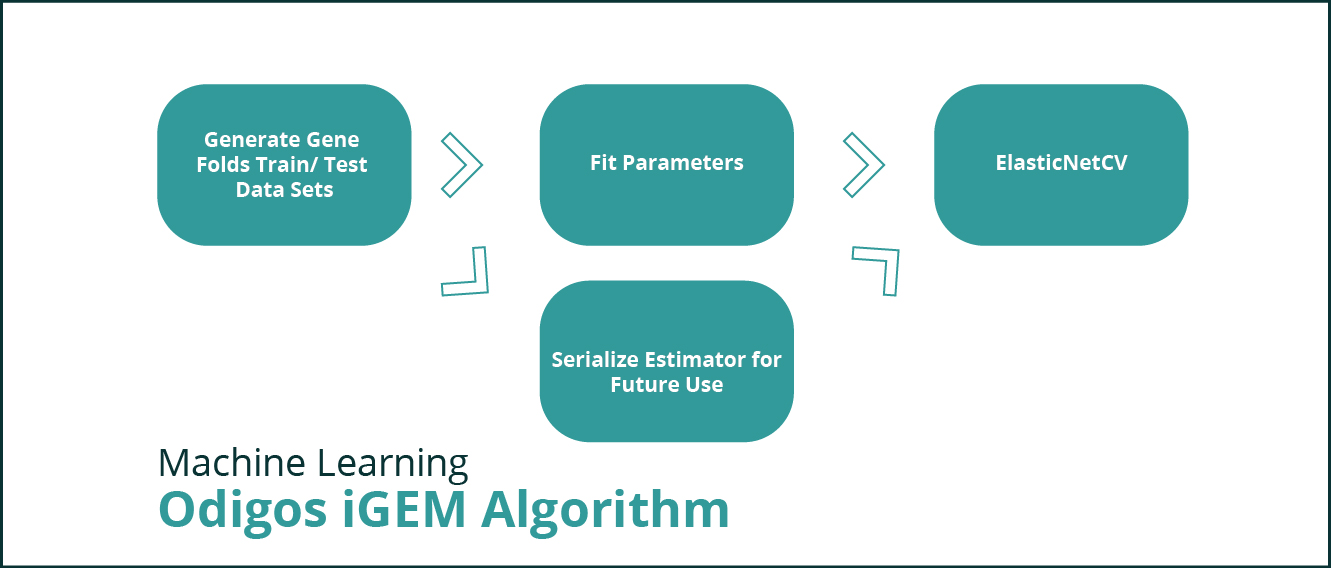
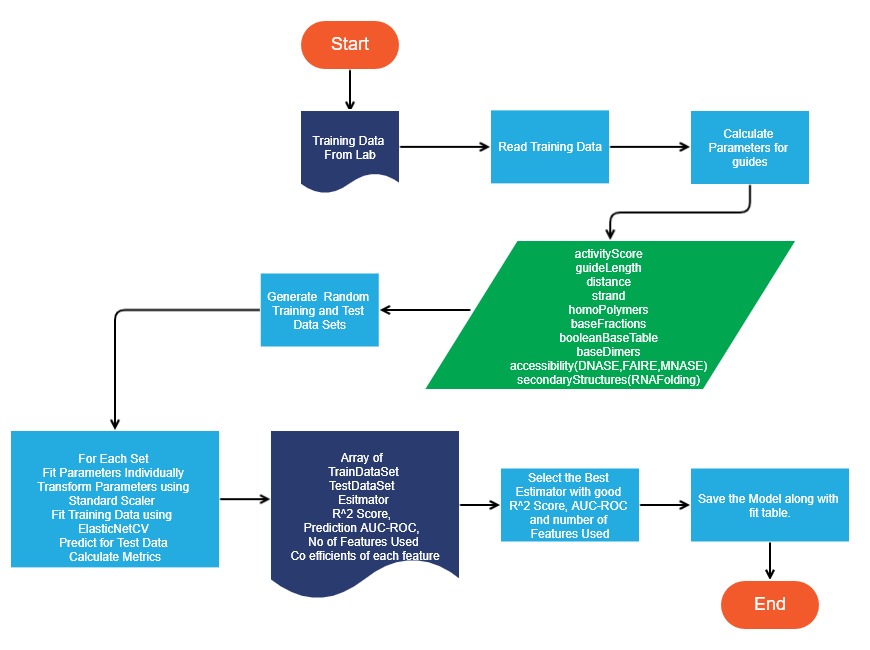
This module is responsible for loading the training data and transforming the parameters in such a way that the data can be fed to a linear regression model. In our application this entry point to this module is a jupyter notebook that can be run in a notebook server. It is iGEM_CRISPRi_Library_Design.ipynb.
This module uses Box Plots and Scatter plots to study parameters and the relationship of those parameters with score.
After training the model, the generated estimator is stored in a folder so that it can be used to do comparative study, score guide RNAs and predict guides for genomic areas of interest.
The following diagram depict the various phases of the module.
This module uses Box Plots and Scatter plots to study parameters and the relationship of those parameters with score.
After training the model, the generated estimator is stored in a folder so that it can be used to do comparative study, score guide RNAs and predict guides for genomic areas of interest.
The following diagram depict the various phases of the module.



Odigos Model Compare
This module is responsible to do the comparative study of the sgRNA scores across Weissman’s algorithm, Odigos iGEM algorithm and calculated in Lab. It first loads the estimator generated by Odigos iGEM Model Generator. Then it finds the guides and calculator scores of them using the estimator.
It calculates the Mean Square Error (MSE) between scores predicted by Weissman’s algorithm and Lab Scores. Similarly, it calculates MSE between Odigos iGEM algorithm scores and Lab Scores.
The comparison of these two MSE values gives us an idea of which score set is closest to lab data.
The entry point to this module in our application is through the jupyter notebook file iGEM_CRISPRi_sgRna_Score_Comparision.ipynb.
It calculates the Mean Square Error (MSE) between scores predicted by Weissman’s algorithm and Lab Scores. Similarly, it calculates MSE between Odigos iGEM algorithm scores and Lab Scores.
The comparison of these two MSE values gives us an idea of which score set is closest to lab data.
The entry point to this module in our application is through the jupyter notebook file iGEM_CRISPRi_sgRna_Score_Comparision.ipynb.
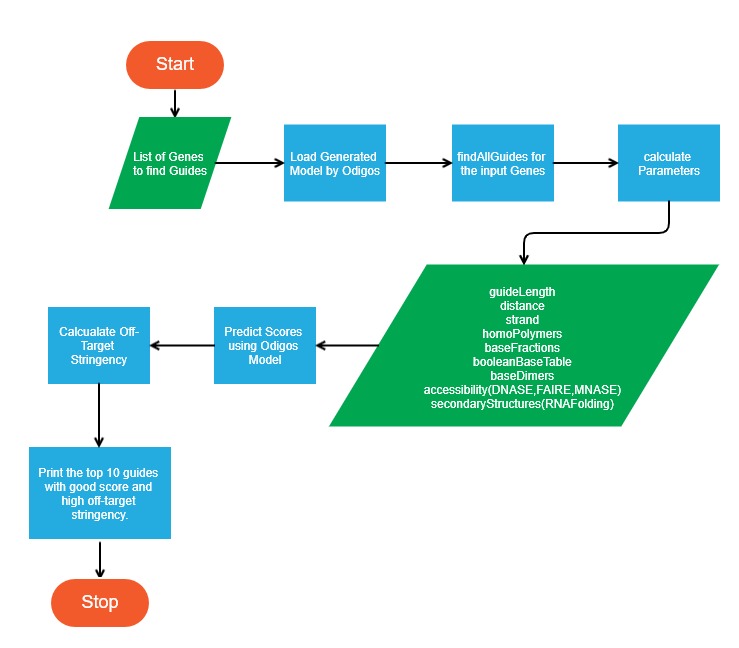
Odigos Guide Predictor
This module is responsible for predicting good guides for the genes that we are interested in. This module loads the estimator generated by Odigos iGEM Model Generator and uses it to predict scores. It allows the end user to specify any gene (Currently from the human genome) as input. Then it uses the genome files and TSS annotations to search for all possible guides. It then uses a trained estimator to create a feature matrix and score the guides.
It then does Off-target-stringency study of the guides and selects the top ten guides for use. Prediction of sgRNA off-target effects is performed using weighted Bowtie (v1.0.0, RRID:SCR_005476 [Ben Langmead et al., 2009]) alignment largely as previously described (Gilbert et al., 2014) with several adjustments. The '--tryhard' flag was added to the Bowtie command to increase sensitivity for mismatched sgRNA target sites.
The entry point to this module in our application is through the jupyter notebook file iGEM_CRISPRi_sgRna_Score_Comparision.ipynb.

Off-target stringency is based on phred-quality score and uses bowtie to calculate quality guides with different thresholds. Following off target levels are considered.
offTargetLevels = [['31_nearTSS', '21_genome'],['31_nearTSS'],['21_genome'],['31_2_nearTSS'],['31_3_nearTSS']]
Highest stringency to lowest are in the following order.
It then does Off-target-stringency study of the guides and selects the top ten guides for use. Prediction of sgRNA off-target effects is performed using weighted Bowtie (v1.0.0, RRID:SCR_005476 [Ben Langmead et al., 2009]) alignment largely as previously described (Gilbert et al., 2014) with several adjustments. The '--tryhard' flag was added to the Bowtie command to increase sensitivity for mismatched sgRNA target sites.
The entry point to this module in our application is through the jupyter notebook file iGEM_CRISPRi_sgRna_Score_Comparision.ipynb.


Off-target stringency is based on phred-quality score and uses bowtie to calculate quality guides with different thresholds. Following off target levels are considered.
offTargetLevels = [['31_nearTSS', '21_genome'],['31_nearTSS'],['21_genome'],['31_2_nearTSS'],['31_3_nearTSS']]
Highest stringency to lowest are in the following order.
- Only one match in 500bp flank region with quality threshold 31 and entire genome with quality threshold 21
- Only one match in the 500bp flanking region
- Only one match in the entire genome with 21 quality threshold value
- Only two matches in the 500bp flanking region with 31 threshold value
- Only three matches in the 500bp flanking region with 31 threshold value
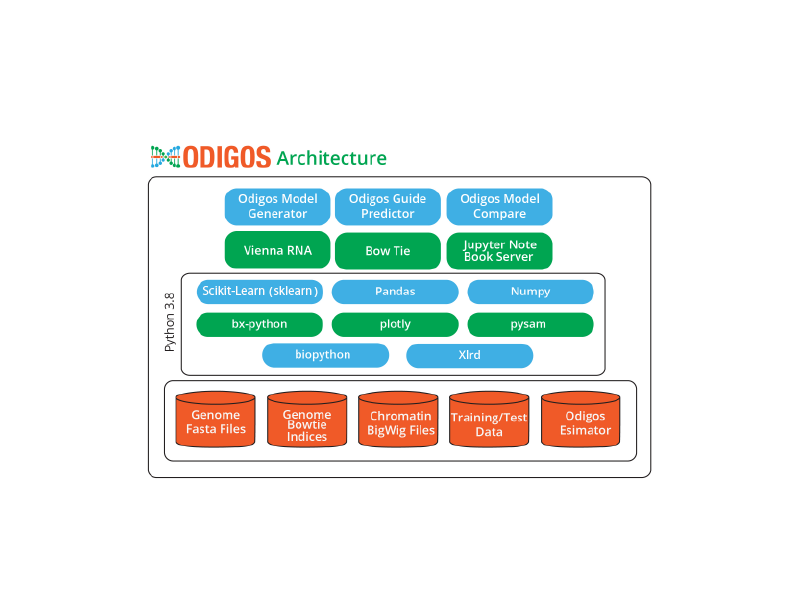
Hardware
It is recommended to use a high end quad-core system with 16GB RAM and 100GB storage and server like Ubuntu 20.0 LTS. The application can run on Mac OS x as well. Currently, Windows has some limitations because few of the libraries required to do some alignment study are missing.
Software
- Python v3.8
- Jupyter notebook
- Biopython
- SciPy/Numpy/Pandas
- Scikit-learn/sklearn
- bx-python (v0.5.0, https://github.com/bxlab/bx-python)
- Pysam
- Plotly
- xlrd
External command line applications required:
- ViennaRNA
- Bowtie (not Bowtie2)
Genomic Data
Large genomic data files required:
- Genome sequence as FASTA (hg19)
- FANTOM5 TSS annotation as BED (TSS_human)
- Chromatin data as BigWig (MNase, DNase, FAIRE-seq)

